Fig. 5
- ID
- ZDB-FIG-210217-53
- Publication
- Begeman et al., 2020 - Decoding an Organ Regeneration Switch by Dissecting Cardiac Regeneration Enhancers
- Other Figures
- All Figure Page
- Back to All Figure Page
|
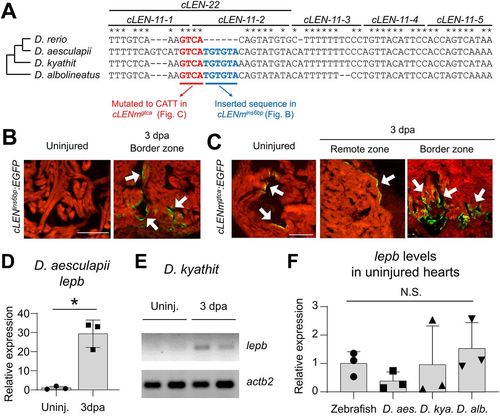
The cLEN repressive element, but not a neighboring sequence, is present in other Danio species. (A) Alignment of orthologous sequences of cLEN11-1 to cLEN11-5 in Danio species, including Danio rerio, Danio aesculapii, Danio kyathit and Danio albolineatus. cLEN-22 contains 6 or 9?bp insertions in non-zebrafish sequences. Asterisks indicate conserved base pairs. The conserved GTCA sequence overlapping cLEN-11-1 and cLEN-11-2 is marked in red. Dendrogram indicates phylogenetic relationship. (B) Images of cardiac sections of cLENins6bp:EGFP transgenic fish harboring a 6?bp insertion (TGTGTA). (C) Images of cardiac sections of cLENmgtca:EGFP transgenic fish harboring the mutation of GTCA to CATT. The arrows indicate endocardial EGFP expression. Numbers of animals are shown in Table S3. (D) RT-qPCR analysis of lepb in uninjured and 3?dpa hearts of D. aesculapii. (E) RT-PCR of samples from the uninjured and 3?dpa hearts of D. kyathit. actb2 was used as the loading control. Uninj., uninjured hearts. (F) RT-qPCR analysis of lepb in the uninjured hearts of zebrafish, D. aesculapii, D. kyathit and D. albolineatus. The data are presented as meanħs.d., n=3; lepb transcript levels were normalized to actb2 levels in D,F. *P<0.05; N.S., not significant. Student's unpaired, two-tailed t-test. Scale bars: 100?µm in B,C. |