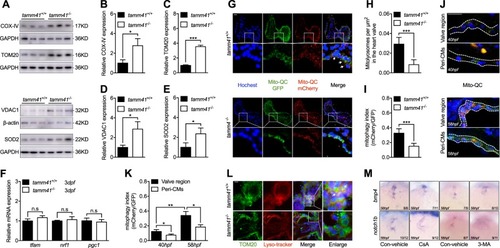
Disturbed mitophagy progression tamm41 deficient embryos. a–eTamm41−/− embryos exhibit increased IMM (COX-IV), OMM (VDAC1, TOM20), and matrix (SOD2) protein levels revealed via western blot analysis. f Normally expressed mitochondrial biogenesis genes (tfam, nrf1, pgc1) in tamm41−/− embryos revealed by qPCR. g–i The mito-QC was used to detect the lysosome-located mitochondria in tamm41+/+ and tamm41−/− hearts. Reduced red fluorescence intensity in tamm41−/− heart valve, which is not observed in the circulating red blood cells. For each group, ten hearts were examined. Scale bar: 20 μm. j, k mito-QC examination of mitophagy activities at 40 hpf and 58 hpf hearts. For each heart (n = 10), a ratio of mCherry to GFP mean fluorescence intensity was generated for the valve or peri-cardiomyocytes (Peri-CMs) region. Scale bar: 20 μm. l Representative images manifest the increased delivery of mitochondria (TOM20, green) to lysosome (lyso-tracker, red) in tamm41+/+ hearts (shown by white arrows), as compared with that in tamm41−/− hearts. For each group, ten hearts were examined. Scale bar: 20 μm. m Reduced heart valve genes, including bmp4 and notch1b, caused by either CsA or 3-MA addition. Black horizontal lines indicate mean ± SD. Means ± SD are shown for three independent experiments. n.s, not significant, *P < 0.05; **P < 0.01; ***P < 0.001 (Student’s t-test)
|

