Fig. 2
- ID
- ZDB-FIG-171108-20
- Publication
- Zhou et al., 2016 - Cardiac sodium channel regulator MOG1 regulates cardiac morphogenesis and rhythm
- Other Figures
- All Figure Page
- Back to All Figure Page
|
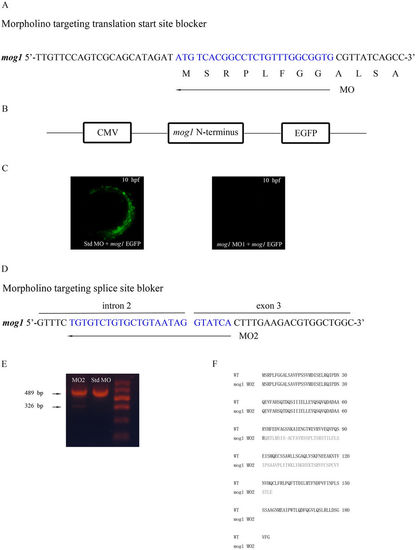
Two mog1 MOs effectively knock expression of mog1 down. (A) Mog1 translation-blocking MO1 was directed against the translation start site. (B) Construction of a pCMV-mog1-EGFP reporter. The 5?-UTR of zebrafish mog1 and a part of the mog1 coding sequence (nucleotide sequences 1?171 starting with A of ATG start codon, amino acid residues 1?57) were fused to the EGFP gene in vector pEGFP-N1, in which expression of the Mog1?EGFP fusion protein (green) is under the control of the CMV promoter. (C) The mog1-EGFP reporter gene was injected into one- to two-cell-stage embryos together with control Std-MO or mog1 MO1. Expression of the Mog1?EGFP fusion protein was examined under a fluorescence microscope at the 10?hpf stage. Note that mog1 MO1 effectively abolished the expression of MOG1?EGFP (lack of green signal) compared with control. (D) Mog1 splicing-blocking MO2 was designed to target the intron 2?exon 3 splicing donor site. (E) Results of RT?PCR analysis with total RNA from 24?hpf embryos treated with MO2 or Std MO. MO2 embryos generated a smaller size of alternatively spliced RT?PCR product (326?bp in MO2 versus 489?bp in Std MO). (F) Sequence analysis of the 326?bp band from MO2-injected embryos revealed that MO2 resulted in an alternatively spliced mog1 transcript with a deletion of exon3 (163?bp). |