FIGURE
Fig. 5
- ID
- ZDB-FIG-090304-3
- Publication
- Slanchev et al., 2009 - Control of Dead end localization and activity – Implications for the function of the protein in antagonizing miRNA function
- Other Figures
- All Figure Page
- Back to All Figure Page
Fig. 5
|
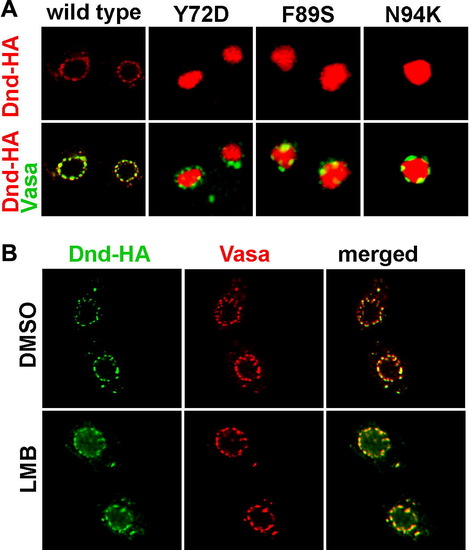
Dead end shuffles between the nucleus and the cytoplasm. (A) Dnd subcellular localization was changed from cytoplasmic to nuclear when amino acids Y72, F89 and N94 were altered. (B) Dead end protein was trapped in the nucleus when the Exportin 1 mediated nucleocytoplasmic export was blocked by leptomycin B (LMB). The localization of Dead end (green) was compared to that of Vasa protein (red), which labels the perinuclear granules in the control embryos treated with DMSO only (upper panel) and LMB treated embryos (lower panel). |
Expression Data
Expression Detail
Antibody Labeling
Phenotype Data
Phenotype Detail
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Reprinted from Mechanisms of Development, 126(3-4), Slanchev, K., Stebler, J., Goudarzi, M., Cojocaru, V., Weidinger, G., and Raz, E., Control of Dead end localization and activity – Implications for the function of the protein in antagonizing miRNA function, 270-277, Copyright (2009) with permission from Elsevier. Full text @ Mech. Dev.