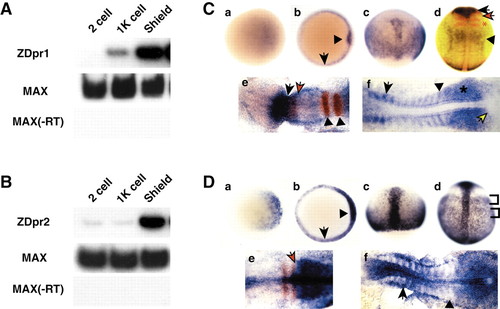
RT-PCR and in situ hybridization analysis of zebrafish dpr1 and dpr2. (A,B) RT-PCR analysis (25 embryos per stage) of dpr1 (A) and dpr2 (B). Zebrafish max is used as a loading/positive control. ?RT is a control for genomic DNA contamination. (C,D) In situ hybridization for expression of dpr1 (C, parts a-f) and dpr2 (D, parts a-f), with embryos shown that are representative of at least 25 analyzed per stage. Stages: (a) sphere stage; (b) shield stage; (c) 80% epiboly; (d) tailbud; (e) five somites (in C), three somites (in D); (f) 12 somites. Orientation: (a,b) animal views, dorsal is rightwards; (c,d) dorsal views, anterior is upwards; (e,f) anterior is leftwards. (C, part b) Arrow indicates margin and arrowhead indicates shield. (C, part d) dpr1 (blue), pax2.1 (red, midbrain/hindbrain boundary) and krox20 (red, ?r3 and 5). Black arrow indicates dpr1 expression in brain. Black arrowhead indicates dpr1 expression in anterior spinal cord. Red arrow indicates pax2.1. Red asterisk indicates krox20. (C, part e) Black arrow is posterior limit of dpr1(blue) expression. Red arrow is posterior limit of pax6.1(red) expression. Arrowheads indicate r3 and r5. (C, part f) Arrow indicates anterior older somite. Arrowhead indicates staining of posterior, younger somite. Asterisk indicates presomitic mesoderm. Yellow arrow indicates tailbud. (D, part b) Arrow indicates margin and arrowhead indicates shield. (D, part d) Brackets indicate mediolateral banding pattern of staining in lateral mesoderm. (D, part e) Arrow indicates boundary of krox20 (red) and dpr2 (blue) staining. (D, part f) Arrow indicates staining in the anterior of the older somites. Arrowhead indicates presomitic mesoderm.
|

