- Title
-
Involvement of Glucosamine 6 Phosphate Isomerase 2 (GNPDA2) Overproduction in β-Amyloid- and Tau P301L-Driven Pathomechanisms
- Authors
- Lachén-Montes, M., Cartas-Cejudo, P., Cortés, A., Anaya-Cubero, E., Peral, E., Ausín, K., Díaz-Peña, R., Fernández-Irigoyen, J., Santamaría, E.
- Source
- Full text @ Biomolecules
|
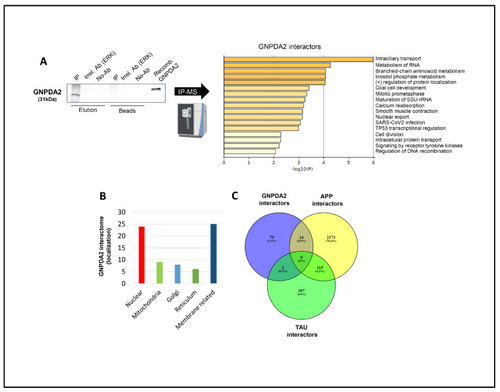
Identification of GNPDA2 molecular interactors. ( |
|
RNA-seq analysis in hNECs. ( |
|
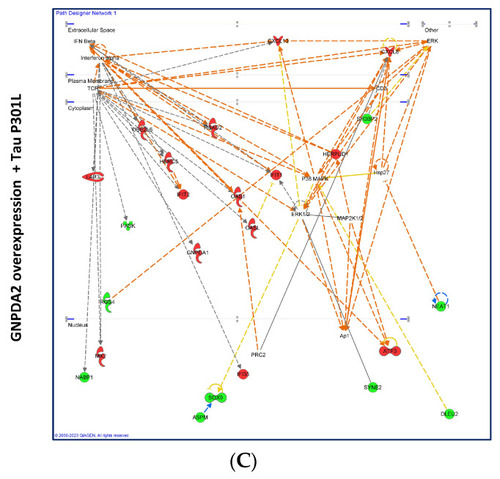
Gene interactome maps for differentially expressed genes in hNECs. Representation of the interactions between differentially expressed genes after GNPDA2 overexpression ( |
|
Gene interactome maps for differentially expressed genes in hNECs. Representation of the interactions between differentially expressed genes after GNPDA2 overexpression ( |
|
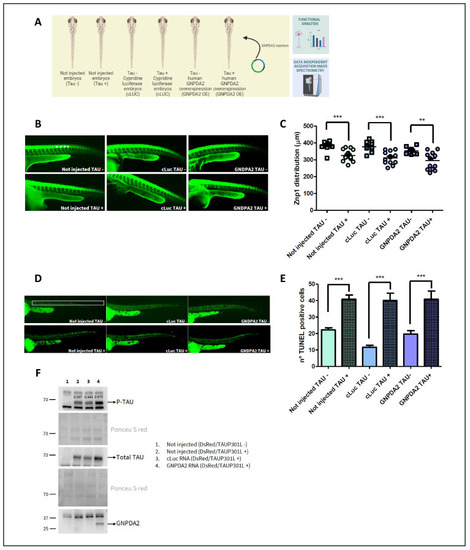
Study of GNPDA2 overexpression in zf Tau P301L transgenic embryos. ( |
|
Proteomics in h.Tau P301L zebrafish embryos. ( |
|
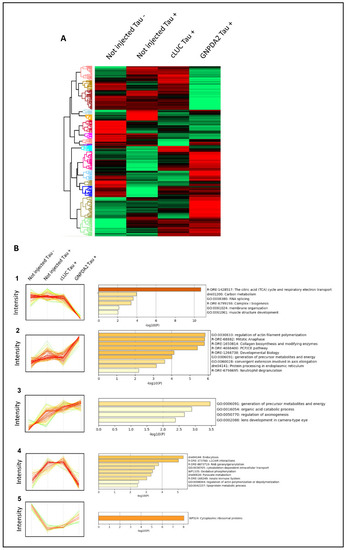
Differentially expressed proteins across Tau P301L embryos overexpressing GNPDA2. ( |
|
|