- Title
-
Disruption of PIKFYVE causes congenital cataract in human and zebrafish
- Authors
- Mei, S., Wu, Y., Wang, Y., Cui, Y., Zhang, M., Zhang, T., Huang, X., Yu, S., Yu, T., Zhao, J.
- Source
- Full text @ Elife
|
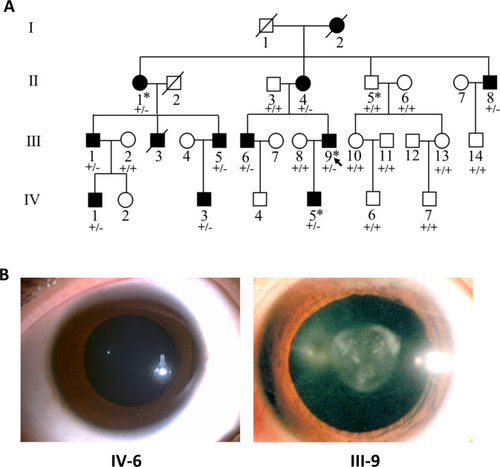
Pedigree structure and ocular manifestations of the cataract family.
(A) Pedigree of the family with congenital cataract. Squares denote males and circles denote females; Symbols crossed by a line indicate deceased individuals. Filled symbols indicate affected individuals, while open symbols indicate unaffected individuals. All affected family members had bilateral congenital cataract. The arrow denotes the proband. The individuals marked with an asterisk (?) are analyzed by whole-exome sequencing. Genotypes of the PIKFYVE variant (p.G1943E) are indicated below each symbol (+, wild-type allele; ?, p.G1943E variant allele). (B) Slit-lamp photographs showing the transparent lens of an unaffected individual (IV-6) and the nuclear pulverulent cataract in the left eye of the proband (III-9). |
|
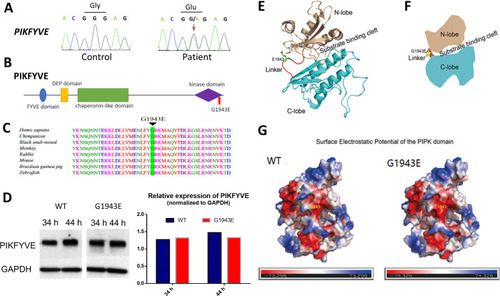
(A) Sanger sequencing chromatogram showing the cDNA sequences from a healthy control and a cataract patient. The heterozygous c.5828G>A missense variant in the patient is indicated by the red arrow. (B) A schematic diagram showing the human PIKFYVE domains. The p.G1943E variant in the PIPK domain is indicated by the red arrow. (C) Protein sequence alignment of PIKFYVE orthologs in vertebrates. The black triangle denotes the conserved glycine at position 1943. (D) Western blot analysis of PIKFYVEWT and PIKFYVEG1943E expression in HEK293T cells that were transiently transfected with either pCS2(+)-CMV-PIKFYVEWT or pCS2(+)-CMV-PIKFYVEG1943E. The protein levels were normalized by GAPDH expression. Experiments were repeated three times. (E) Predicted structure model of the p.G1943E variant form of PIKFYVE PIPK domain generated by the PHYPRE2 server (http://www.sbg.bio.ic.ac.uk/~phyre2/html/). N-lobe, C-lobe, and the hinge linker are shown in gold, cyan, and red, respectively. The variant residue E1943 is shown in sticks and labeled with green. The negatively charged residue D1872 close to E1943 side chain is also shown in sticks. (F) A schematic demonstrating the organization of PIKFYVE PIPK domain. N-lobe, C-lobe, and the hinge linker are shown in gold, cyan, and red, respectively. The position of the p.G1943E variant is labeled with a yellow star. (G) Surface electrostatic potential comparison of the PIPK domain of PIKFYVE between wild-type (WT) and p.G1943E variant. The electrostatic potentials are presented as heatmaps from red to blue, and the electrostatic potential scales are shown in the lower panel. See Figure 2?source data 1 for details. |
|
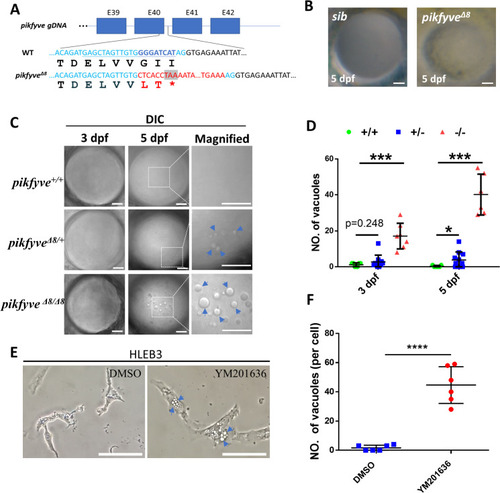
(A) A schematic diagram showing the generated pikfyve?8 mutant allele. The underlined base pairs are the sgRNA target. The deleted base pairs are shown in dark blue while inserted ones are shown in red. The stop codon introduced in the mutant form is shown in the grey box. (B) Representative images showing the lens of sibling and pikfyve?8 mutants at 5 dpf. (C) Representative differential interference contrast (DIC) images showing the lens of pikfyve+/+, pikfyve+/?8, and pikfyve?8/?8 embryos at 3 dpf and 5 dpf. The scale bars represent 10 ?m in (B) and (C). (D) Quantification of vacuole number in the lens of pikfyve+/+, pikfyve+/?8, and pikfyve?8/?8 embryos at 3 dpf (n=7 for pikfyve+/+; n=10 for pikfyve+/?8; n=7 for pikfyve?8/?8) and 5 dpf (n=7 for pikfyve+/+; n=11 for pikfyve+/?8; n=6 for pikfyve?8/?8). (E) Representative images of HLEB3 cells treated with DMSO or PIKFYVE inhibitor YM201636 for 4 hr. The scale bars represent 25 ?m. (F) Quantification of the vacuole numbers in (E). ****, p<0.0001, Student?s t-test. All experiments were repeated three times. See Figure 3?source data 1 for details.
|
|
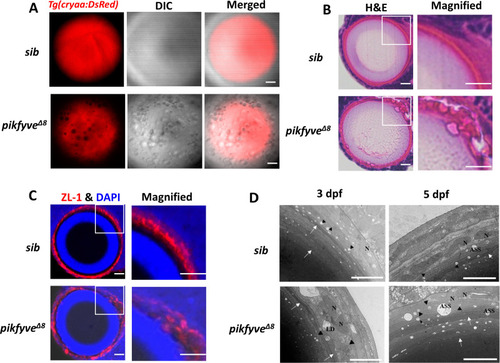
(A) Confocal imaging of the lens of 5-dpf siblings and pikfyve?8 mutants in Tg(cryaa:DsRed) transgenic background. (B) Hematoxylin-eosin (HE) staining of 5-dpf siblings and pikfyve?8 mutant zebrafish lens after cryostat section. (C) ZL-1 antibody and DAPI staining of 5-dpf siblings and pikfyve?8 mutant zebrafish lens. (D) Transmission electron microscope (TEM) images of the lens of siblings and pikfyve?8 mutants at 3 dpf and 5 dpf. ASS, autophagy lysosome; LD, lipid droplet; N, nucleus. All results were confirmed in three different individuals. All the scale bars represent 10 ?m. |
|
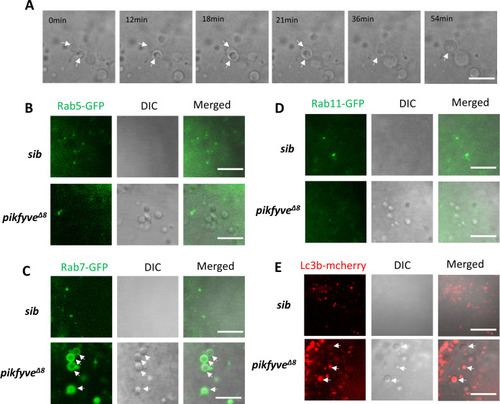
(A) Time-lapse imaging indicating the dynamic changes of vacuole formation in the lens of 4-dpf pikfyve?8 mutants. White arrows indicate the fusion process of two small vacuoles. (B) Representative images showing the lens of 3.5-dpf siblings and pikfyve?8 mutants injected with gfp-rab5c mRNA. (C) Representative images showing lens of 3.5-dpf siblings and pikfyve?8 mutants injected with gfp-rab7 mRNA. (D) Representative images showing lens of 3.5-dpf siblings and pikfyve?8 mutants injected with gfp-rab11a mRNA. (E) Representative images showing lens of 3.5-dpf siblings and pikfyve?8 mutants injected with mcherry-lc3b mRNA. All experiments were repeated three times. All the scale bars represent 10 ?m. |
|
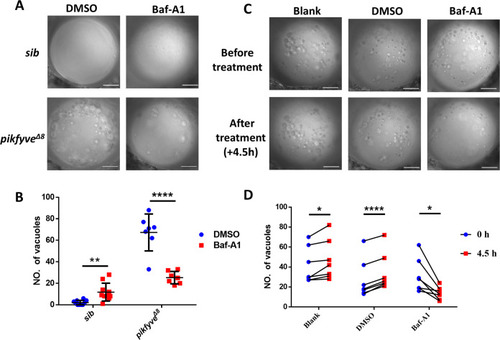
(A) Representative confocal images of the lens of 4-dpf pikfyve?8 mutants treated with DMSO or Baf-A1 for 4.5 hr. (B) Quantification of the vacuole numbers in the lens of 4-dpf siblings and pikfyve?8 mutant embryos treated with DMSO or Baf-A1 (n=10 for sibling groups; n=7 for mutant groups). (C) Confocal images of the lens of 4-dpf pikfyve?8 mutants with no treatment or after 4.5 hr treatment with DMSO or Baf-A1. (D) Quantification of the vacuole numbers in (C) (n=6 for each group). All experiments were repeated three times. All the scale bars represent 20 ?m. *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001, Student?s t-test. See Figure 6?source data 1 for details.
|