- Title
-
A missense mutation in SNRPE linked to non-syndromal microcephaly interferes with U snRNP assembly and pre-mRNA splicing
- Authors
- Chen, T., Zhang, B., Ziegenhals, T., Prusty, A.B., Fröhler, S., Grimm, C., Hu, Y., Schaefke, B., Fang, L., Zhang, M., Kraemer, N., Kaindl, A.M., Fischer, U., Chen, W.
- Source
- Full text @ PLoS Genet.
|
(A), Family pedigree. Filled symbol indicates individual suffering from non-syndromal primary microcephaly and intellectual disability. (B), Traditional Sanger sequencing validated the identified |
|
(A-E), The NSM mutation in SmE impairs its interaction with U snRNP assembly machinery and incorporation into U snRNPs. (A), Anti-FLAG immunoprecipitation after transient transfection in HEK293T cells and western blotting analysis for co-precipitated U snRNP intermediates. Mock immunoprecipitations were performed with untransfected lysates. (B), 3’-end labeling of co-precipitated RNA and autoradiography. RNA immunoprecipitated using the H20 antibody against m3G/m7G cap of U snRNAs was used as reference. (C), Quantification of the data shown in A and B from two independent biological replicates, with black bars representing wild type and gray bars representing mutant SmE. (D), Predicted structural model for interference of the SmE mutation in its interaction with Gemin2, based on the PDB structure 4V98. (E), Immunoprecipitation using antibodies specific to Sm proteins, SMN, pICn and U snRNA cap, with lysates from HEK293T cells transfected with dual expression plasmid encoding 2A-tagged mutant SmE and HA-tagged wild type SmE and western blotting to analyze the integration of the wild type and mutant SmE into U snRNP biogenesis pathway. |
|
(A-B), Indirect immunofluorescence and confocal microscopy of HeLa cells transfected with either FLAG-tagged wild type or mutant SmE (WT/Mut) or left untransfected (negative control). Empty white arrowheads indicate localization pattern observed and filled white arrowheads indicate zoomed in region shown in the overly inset. (A), Top panel shows clear co-localization of wild type SmE (green) and coilin (magenta) in CBs and middle panel shows most of the mutant SmE (green) distributed in the cytoplasm with a minor fraction in the nucleus and co-localizing with coilin (magenta) in CBs. (B), While wild type SmE (green, top panel) co-localizes with SmD3 (magenta) in CBs and splicing speckles, the mutant SmE (green, middle panel) is predominantly cytoplasmic with marginal co-localization with SmD3 in CBs or in nuclear speckles. |
|
(A), The mRNA splicing in patient derived fibroblast cells is impaired. MA plot shows the intron retention was dramatically increased in patient derived fibroblast cells. X axis: the sum of log2 transformed splicing in and splicing out reads number for each intron. Y axis: difference in percentage of intron retention (PIR) between fibroblast derived from the patient (mutant) and healthy control. (B), The intron retention leads to decreased gene expression. Backgrounds are those genes without any intron showing significantly increased retention. (C), Heatmap illustrating expression of 11670 protein coding genes (average RPKM>1) in HEK293 cells among different experimental conditions. Control, control siRNA; KD, SmE siRNA; KD+WT, SmE siRNA+wild type SmE; KD+Mut, SmE siRNA+mutant SmE. (D), Number of aberrant splicing events induced by SNRPE/SmE knockdown (KD) comparing with control. RI, retained intron; SE, skipped exon; MXE, mutually exclusive exon; ASS, alternative splice site. (E), Numbers of aberrant splicing events in KD, KD+Mut and KD+WT comparing to control. (F), The intron retention leads to decreased gene expression. Backgrounds are those genes without any intron showing significant increased retention. |
|
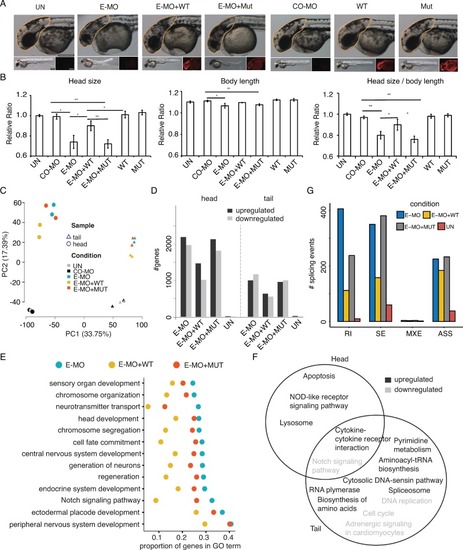
(A), Measurement of zebrafish head size across different experimental conditions. CO-MO, control morpholino; E-MO, SmE morpholino. The morpholino and/or PHENOTYPE:
|
|
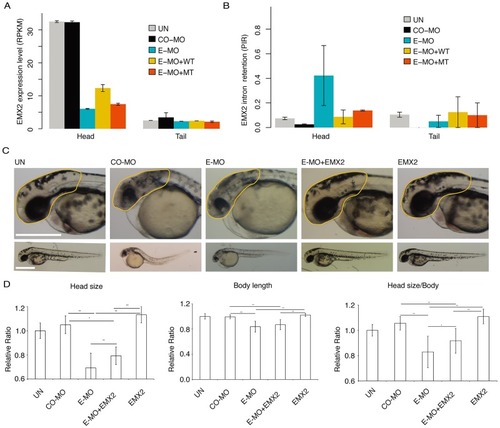
(A), The expression of |