- Title
-
Corepressor diversification by alternative mRNA splicing is species specific
- Authors
- Privalsky, M.L., Snyder, C.A., Goodson, M.L.
- Source
- Full text @ BMC Evol. Biol.
|
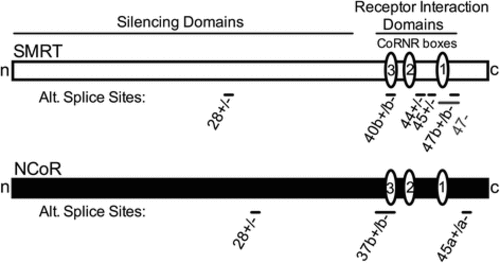
Schematic of alternative splice sites in SMRT and NCoR. The longest known corepressor alternative splice variants are depicted as horizontal rectangles. Alternative splice locations are shown as bars and exon numbers are indicated as in RefSeq. Interaction domains for additional components of the corepressor complex (?silencing domains?) and for nuclear hormone receptors (?receptor interaction domains? containing the CoRNR box motifs, represented as ovals) are shown |
|
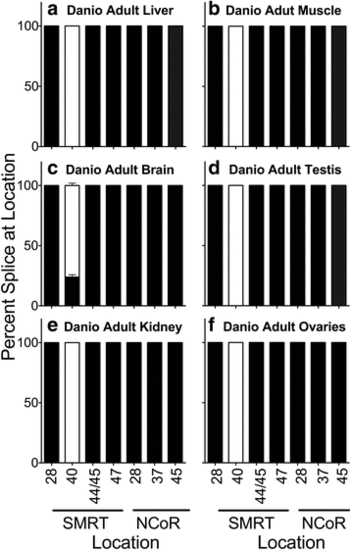
Alternative splicing in adult Danio rerio. The organs indicated were isolated, RNA was extracted, and the alternative splicing at the indicated locations was analyzed by use of suitable primers and RT-PCR as in Methods. The percentage of each alternatively spliced corepressor variant produced at each location was quantified by scanning and is presented as stacked bars adding to 100 %. Mean and standard deviation are shown (n?=?5) for each. Black filled bars represent the longest splice variant at each location and open fill represents the shortest splice variant at each location. Panels represent Danio adult liver (a), Danio adult muscle (b), Dano adult brain (c), Danio adult testis (d), Danio adult kidney (e), and Danio adult ovaries (f) |
|
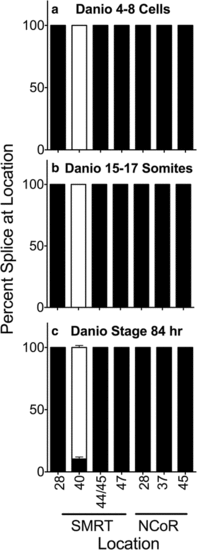
Alternative splicing during Danio rerio development. Entire individuals were isolated at the stages indicated, RNA was extracted, and the alternative splicing at the indicated locations was analyzed and is presented graphically as in Fig. 2. Mean and standard deviation are shown (n?=?3) for each. Panels represent Danio 4-8 cell stage animals (a), Danio 15-17 somite stage animals (b) and Danio stage 84 hour animals (c) EXPRESSION / LABELING:
|
|
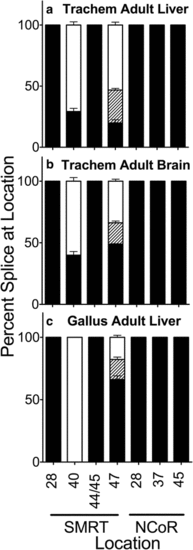
Alternative splicing in Trachemys scripta liver and brain, and in Gallus gallus adult liver. Adult Trachemys liver (a), Trachemys brain (b), or Gallus liver (c) were isolated as indicated, RNA was extracted, and the alternative splicing at the indicated locations was analyzed and is presented graphically as in Fig. 2; the SMRT exon 47b- variant, of intermediate length between the 47b?+?and 47- variants, is indicated by diagonal striping. Mean and standard deviation are shown (n???3) for each |
|
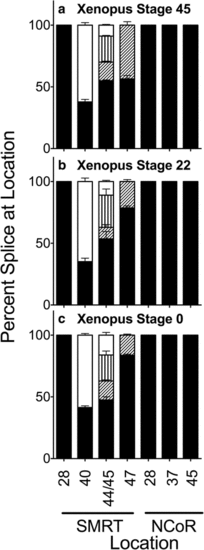
Alternative splicing in whole Xenopus at stages indicated. RNA was isolated from entire Xenopus individuals at the stages denoted and the alternative splicing at the indicated locations was analyzed and is presented graphically as in Fig. 2. Fill patterns are as in Figs. 2 and 4 with one exception: four variants are generated from the S44 and S45 positions in this species (S44+/45+. S44+/45-, S44-/45+, and S44-/S45- from largest to smallest; represented by black, diagonal stripe, vertical stripe, and open fill respectively). Mean and standard deviation are shown (n?=?3) for each |
|
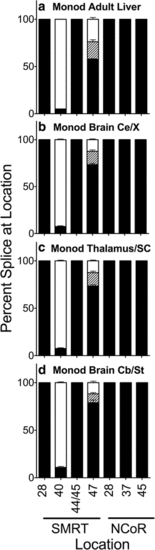
Alternative splicing in adult Monodelphis domestica (opossum). Liver (a) or the parts of the brain indicated (cerebral cortex (b), thalamus/superior colliculus (c), and cerebellum/brain stem(d)) were isolated by dissection, RNA was extracted, and alternative splicing at the indicated locations was analyzed and is presented graphically as in Fig. 2. Mean and standard deviation are shown (n?=?3) for each |
|
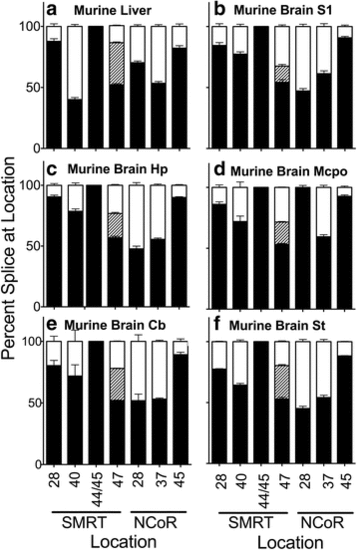
Alternative splicing in adult Mus mus domestica (mouse). Liver (a) or the parts of the brain indicated: S1 cerebral cortex (b), hippocampus (c), magnocellular preoptic (d), cerebellum (e), and brain stem (f), were isolated by dissection. RNA was extracted and the alternative splicing at the indicated locations was analyzed and is presented graphically as in Figs. 2 and 4. Mean and standard deviation are shown (n?=?3) for each |
|
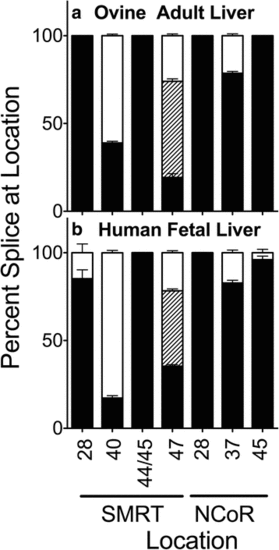
Alternative splicing in ovine and human liver. Livers were isolated, RNA was extracted, and the alternative splicing at the indicated locations was analyzed and is presented graphically as in Figs. 2 and 4. Mean and standard deviation are shown (n?=?3) for each. Panels represent ovine adult liver (a) and human fetal liver (b) |
|
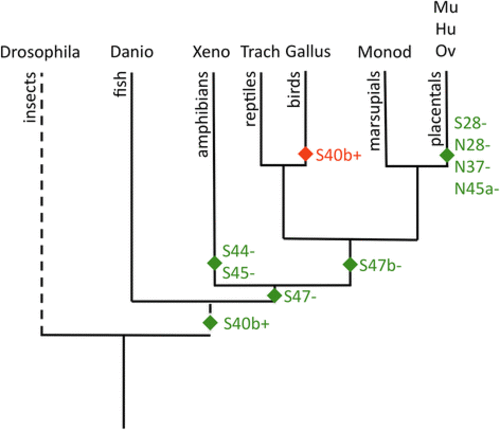
Evolution of alternative splicing for SMRT and NCoR. A dendrogram representation of evolutionary divergence is presented for the species analyzed at the alternatve splice sites previously reported. Dashed lines indicates use of different time scale for the Drosophila divergence. The postulated gain (green symbols) or loss (red symbol) of each alternative splicing site is depicted near the base of the relevant lineages (S?=?SMRT and N?=?NCoR]. A tissue-specific dendrogram would differ only by the absence of detectable S40 alternative splicing in Danio tissues other than brain and whole 84 h. hatchlings; otherwise the different tissues within a given species vary in the abundance of the alternative spliced variants noted here, but not in their presence or absence |