- Title
-
Transcriptional components of anteroposterior positional information during zebrafish fin regeneration
- Authors
- Nachtrab, G., Kikuchi, K., Tornini, V.A., and Poss, K.D.
- Source
- Full text @ Development
|
Regeneration of pattern and underlying differential gene expression in pectoral fins. (A) Zebrafish pectoral fins possess and regenerate an anteroposterior (AP) skeletal pattern. Early osteoblasts marked by osx:EGFP-CAAX (green) are present in the regenerate prior to the onset of AP differences, which arise between 5 and 7 days post-amputation (dpa). Arrowheads indicate amputation plane. BF, bright field. (B) Zebrafish robustly regenerate the AP bone pattern. Quantification of the relative ratios between the width of the uninjured portion of anterior ray 3 (blue) and the widths of regenerating rays across the AP axis. n=12; bar indicates mean. (C) Diagram of a pectoral fin defining the two regions used for RNA-Seq (circled AP1 and AP5) and the five regions (AP1-AP5) used for subsequent qPCR validations. (D) Each AP region of uninjured zebrafish pectoral fins expresses a unique AP code of patterning transcription factor genes. Results shown reflect qPCR confirmation of RNA-Seq data, normalized to β-actin 1 (actb1) levels. n=3. (E) The AP regionalized expression of transcription factor genes is maintained during regeneration. qPCR expression profiles at 4 dpa, normalized to actb1 levels. n=3; mean ± s.e.m.; *P<0.05, **P<0.005, Student?s t-test. |
|
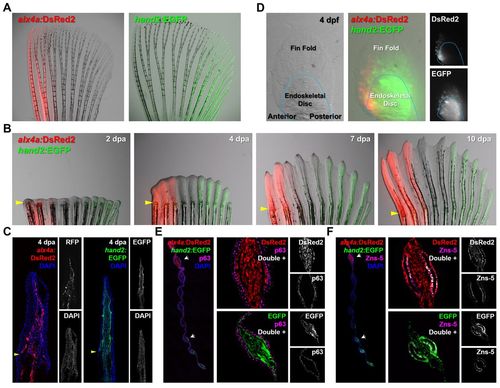
Visualization of AP region-specific transcription factor expression in fin fibroblasts and osteoblasts. (A) Expression of fluorescent transgenic reporters in adult pectoral fins. The alx4a:DsRed2 domain ranges from the most anterior to the third ray. The hand2:EGFP domain extends from the posterior edge to the sixth ray of the fin. (B) The AP expression characteristics of alx4a:DsRed2 and hand2:EGFP are maintained throughout regeneration. Arrowheads indicate amputation plane. (C) Longitudinal sections at 4 dpa confirm reporter expression to nearly the distal tip of the fin. Arrowheads indicate amputation plane. (D) Expression of fluorescent reporters in embryonic pectoral fins. At 4 days post-fertilization (dpf), both reporters display region-specific expression. The expression is similar to that of the adult, but the double-negative medial region is not yet defined. (E) Transverse sections indicate that the expression of each reporter is restricted to the fin mesenchyme. The antibody against p63 marks fin epidermis adjacent to the mesenchymal compartment. Arrowheads indicate amputation plane. (F) Transverse sections indicating that both alx4a:DsRed2 and hand2:EGFP are expressed in a population of fin osteoblasts identifiable by Zns-5 immunoreactivity. Arrowheads indicate amputation plane. |
|
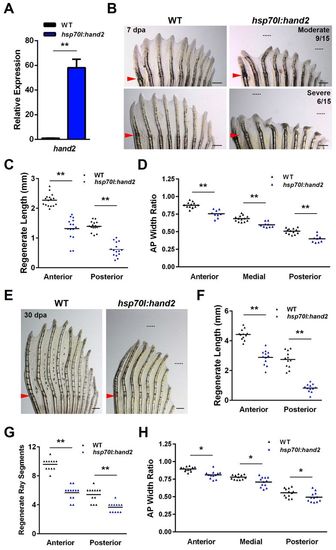
Overexpression of hand2 during fin regeneration alters ray patterning. (A) hand2 expression is induced 58-fold in anterior regions of hsp70l:hand2 pectoral fins 4 hours after a single heat shock at 4 dpa. Values are normalized to actb1 levels and relative to wild-type controls. n=3; mean ± s.e.m. (B) Appearance of hsp70l:hand2 and wild-type clutchmate fins at 7 dpa after a series of daily heat shocks. hand2 overexpression generates shorter rays with a reduced number of bone segments. Phenotypes range from moderate (upper right) to severe (lower right). Representative regenerative growths of wild-type rays 3 and 8 are denoted by dotted lines. (C) Overexpression of hand2 reduces the lengths of regenerating rays across the AP axis of pectoral fins. n=16 (wild type) and n=15 (hsp70l:hand2). (D) Overexpression of hand2 during regeneration reduces the widths of regenerating fin rays across the AP axis. Transgenic fish that displayed a moderate phenotype were quantified. n=13 (wild type) and n=6 (hsp70l:hand2). (E) Appearance of hsp70l:hand2 and wild-type clutchmate fins at 30 dpa after daily heat shocks. hsp70l:hand2 regenerates remain stunted with shorter and thinner fin rays. Representative regenerative growths of wild-type rays 3 and 8 are denoted by dotted lines. (F) Quantification of hsp70l:hand2 ray lengths at 30 dpa. n=12. (G) Quantification of hsp70l:hand2 segment numbers at 30 dpa. n=12. (H) Quantification of hsp70l:hand2 ray widths at 30 dpa. n=12. *P<0.05, **P<0.005, Student?s t-test; bar indicates mean. Arrowheads (B,E) indicate amputation plane. Scale bars: 0.5 mm. |
|
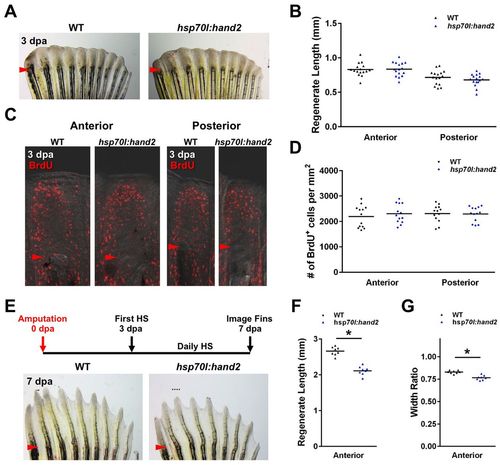
hand2 overexpression exerts its effects during later stages of regeneration. (A,B) No difference in the appearance of hsp70l:hand2 and wild-type clutchmate fins at 3 dpa after a series of daily heat shocks. n=16. (C,D) Blastemal proliferation as measured by BrdU incorporation is also unaffected by hand2 overexpression. n=13. (E) Stunted regeneration in an hsp70l:hand2 pectoral fin compared with a wild-type clutchmate fin at 7 dpa after a series of daily heat shocks beginning at 3 dpa. Representative regenerative growth of wild-type ray 3 is denoted by a dotted line. (F,G) Significant reductions in anterior ray lengths and widths manifest when hand2 is overexpressed during later stages of regeneration. n=8. *P<0.05, Student?s t-test; bar indicates mean. Arrowheads (A,C,E) indicate amputation plane. |
|
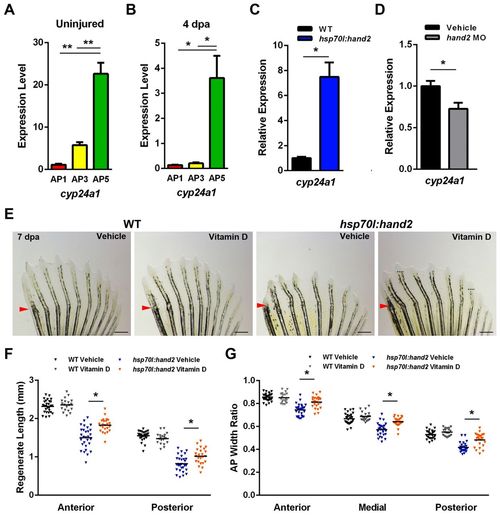
AP regionalization of vitamin D signaling controlled by Hand2. (A) cyp24a1, which encodes a vitamin D-inactivating enzyme, is more highly expressed in posterior regions of uninjured pectoral fins. (B) Posterior enrichment of cyp24a1 is maintained during regeneration at 4 dpa. (C) hand2 induction daily during regeneration elevates anterior cyp24a1 expression <7.5-fold at 4 dpa relative to heat-shocked wild-type clutchmate controls. (D) hand2 morpholino injection into pectoral fin regenerates reduces posterior cyp24a1 expression by <28% relative to vehicle-injected clutchmate controls. (A-D) Expression is normalized to actb1. *P<0.05, **P<0.005, Student?s t-test. n=3; mean ± s.e.m. (E) Appearance of 7-dpa fins from hsp70l:hand2 and wild-type clutchmates given a daily heat shock and either daily vehicle or vitamin D injections. Vitamin D injection had little effect on wild-type regeneration, but partially suppressed the effects on bone patterning caused by hand2 overexpression. Representative regenerative growth of hsp70l:hand2 vehicle-injected rays 3 and 8 is denoted by dotted lines. Arrowheads indicate amputation plane. (F) Quantification of the effects of daily vitamin D injection on ray lengths, indicating improvement in hsp70l:hand2 animals. (G) Daily vitamin D injection increases ray widths in regenerating hsp70l:hand2 fins. (F,G) n=20-28; bar indicates mean. **P<0.05, Student?s t-test. Scale bars: 0.6 mm. |
|
Model for Hand2 influences on positional memory. Adult zebrafish pectoral fins maintain region-specific expression of transcription factor genes. Levels of the posterior transcription factor gene hand2 can help control bone patterning during pectoral fin regeneration. Hand2 levels regulate bone formation during regeneration by direct or indirect regulation of the vitamin D-inactivating enzyme encoded by cyp24a1, restricting bone formation in the posterior rays of pectoral fins. |
|
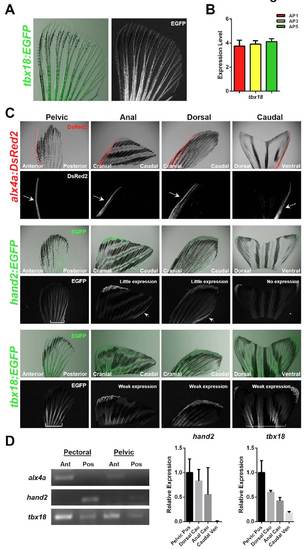
Developmental patterning transcription factor expression domains in adult zebrafish fins. (A) Expression of the tbx18:EGFP fluorescent transgenic reporter across the entire AP axis of an adult pectoral fin. (B) Relatively uniform expression of tbx18 is verified by qPCR. Expression is normalized to actb1. n=3. (C) Expression of alx4a:DsRed2, hand2:EGFP and tbx18:EGFP in the indicated adult fin types. Arrows or brackets indicate expression domains. (D) Endogenous transcription factor gene expression is consistent with BAC reporter fluorescence patterns. (Left) RT-PCR supports region-specific expression of alx4a and hand2 in uninjured paired fins. (Right) qPCR indicates decreased expression of hand2 and tbx18 in all uninjured medial fins when compared with paired fins. Expression is normalized to actb1, relative to levels in posterior regions of pelvic fin. n=2. |
|
Anterior transcription factor overexpression has little or no effect on pectoral fin regeneration. (A) Expression of alx4a, id4 or lhx9 is induced in posterior pectoral fin regions at 10-fold or higher levels in new transgenic lines, 4 hours after a single heat-shock at 4 dpa. Values are normalized to actb1 levels and relative to wild-type controls. n=3; **P<0.005, Student?s t-test; mean ± s.e.m. (B) Appearance of transgenic and wild-type clutchmate fins at 7 dpa after a series of daily heat shocks. Transgenic fins appear similar to wild-type fins. (C) Quantification of regenerate lengths indicates a minor change in some rays after anterior gene overexpression. n=12-16; *P<0.05, Student?s t-test. (D) Quantification of width ratios indicates a minor change in some rays. n=12-16; *P<0.05, Student?s t-test. Scale bars: 0.5 mm. |