- Title
-
A role for N-cadherin in mesodermal morphogenesis during gastrulation
- Authors
- Warga, R.M., and Kane, D.A.
- Source
- Full text @ Dev. Biol.
|
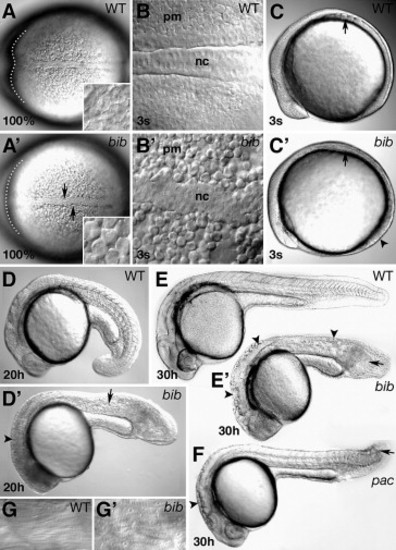
Morphology of the bib phenotype. (A) Dorsal view; arrows indicate kinked mutant notochord, inset shows high magnification of rounded cells in paraxial mesoderm, dotted line outlines ventral portion of anterior neural plate. (B) High magnification of notochord (nc) and paraxial mesoderm (pm). (C) Side view: arrows indicate position of somite furrows; arrowhead indicates flatter mutant tailbud. (D) Side view: arrowhead indicates defects in brain subdivisions; arrow indicates defects in muscle segmentation. Side views of (E) wild-type and bib siblings and (F) pac mutant; arrowheads indicate uneven surface of neural tube, arrows indicate vacuoles in disorganized tail. (G) High magnification of wild type and bib striated muscle. PHENOTYPE:
|
|
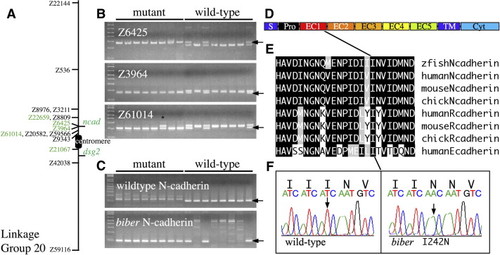
Identification of bib as N-cadherin. (A) Genetic map of Linkage Group 20. Shown are the positions of microsatellite (Z) markers used in mapping and the genes N-cadherin (ncad) and desmoglein2 (dsg2). Markers in green were used to calculate recombination frequencies in Table 1. (B) Examples of products obtained for three closely linked markers. Arrows indicate product that segregates with mutant allele; black asterisk denotes a homozygous mutant individual possessing both products indicating a recombination event between the marker and the N-cadherin locus. (C) Products obtained from same embryos shown in panel B using the wild-type or mutant N-cadherin-specific primers. The expected product (arrow) is 200 bp. (D) Domain structure of the N-cadherin protein. Cyt, cytoplasmic domain; EC1?5, extracellular domains; Pro, prodomain; S, signal sequence; TM, transmembrane domain. (E) The zebrafish amino acid sequence for the carboxyl-end of EC1 containing the bib mutation and the corresponding amino acid sequence alignments for human, mouse and chick and more distantly related cadherins. Identical residues, black; conserved residues, grey. (F) Sequence trace data showing site of mutation. |
|
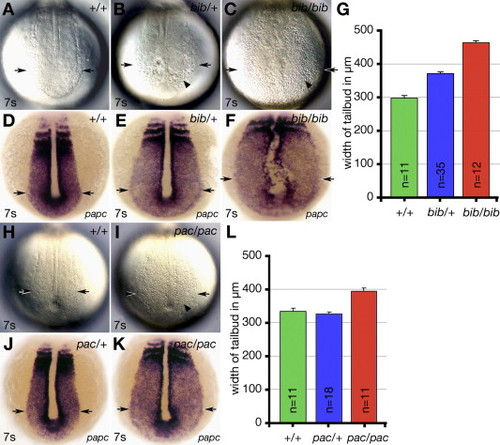
bib has a semi-dominant phenotype. (A?F and H?K) Vegetal views, showing cellular morphology and width of tailbud in (A?F) bib siblings and (H?K) pac siblings. (A?C and H?I) Live embryos and (D?F and J?K) papc expression. Arrows indicate width of tailbud and arrowheads indicate patches of round disaggregated cells. (G and L) Comparison of width of tailbud using papc expression based on genotype of embryos. EXPRESSION / LABELING:
PHENOTYPE:
|
|
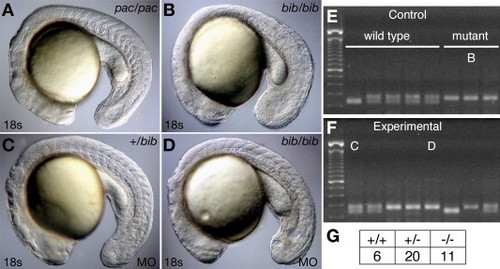
Antisense knockdown of the bib mutant product phenocopies the pac mutant. (A) Homozygous pac mutant, (B) uninjected homozygous bib mutant, and (C) heterozygous and (D) homozygous bib mutant MO-injected siblings showing phenocopy of pac homozygote. Examples of PCR products from the closely linked Z6425 marker used for genotyping (E) uninjected controls and (F) MO-injected experiments; individuals B?D are indicated. (G) The ratio of genotypes in the MO experiment is not statistically different from the expected 1:2:1 ratio. PHENOTYPE:
|
|
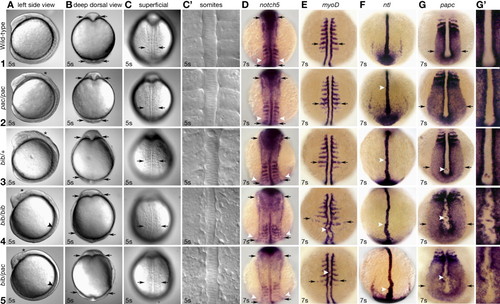
Comparison of the bib and pac phenotypes. Rows (1?5) indicate genotype of the individual. In the case of all bib and wild-type individuals, all which are siblings, this was determined by linkage mapping, with the exception of ntl and notch5 expression, where the genotype was inferred by phenotype. Also, for the pac homozygotes and bib/pac transheterozygotes, genotype was based on phenotype. Columns (A?C) show the same live embryo from a different view. Columns (D?G) show whole-mount in situ hybridizations. (A) Asterisk indicates uneven neural keel; arrowhead indicates absence of somite furrows. (B) Optical cross section at level of midbrain?hindbrain, arrows indicate width of neural plate. (C) Superficial view of somite region, arrows indicate width of paraxial mesoderm at level of 4th somite. (C2) Magnified view showing epithelial organization. (D) Dorsal view, arrows indicate width of neural plate; arrowheads indicate width of paraxial mesoderm. (E) Dorsal view, arrows indicate width of paraxial mesoderm, arrowhead indicates cells in notochord domain. (F) Dorsal view, arrowhead indicates knobby notochord protrusions. (G) Vegetal view, arrows indicate width of tailbud, arrowhead indicates cells in notochord domain. (G′) Magnified view showing irregular paraxial?notochord border and ectopic cells. EXPRESSION / LABELING:
PHENOTYPE:
|
|
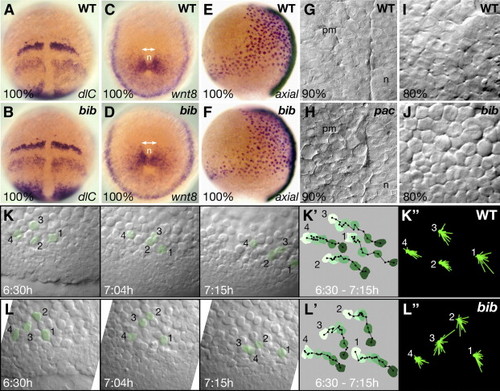
Abnormal cell behavior of the mesoderm in pac and bib mutant gastrulas. (A?D) Whole-mount in situ hybridizations of sibling embryos, double-headed arrows indicate broadened mesodermal territories in the mutant. (E, F) Comparison of axial expression shows normal width of endoderm in mutants. (G?J) High magnification view of the mesodermal layer in live embryos. (G?H) In pac siblings, view of the paraxial mesodermal (pm) territory adjacent to the notochord (n). (I?J) In bib siblings, view of the paraxial mesodermal territory 45° distant from the midline. (K, L) Frames from two simultaneous video timelaspes over a 45-min time period. (K′, L′) Tracings of the net movement for the indicated cells shown in panels K and L; each color is the filled outline of that particular cell for a specific segment in the time-lapse in which time is reflected by change from light green to dark green, filled black circles are the position of the nucleus in that particular cell at 3-min time intervals, black lines are the individual vectors from time point to time point. (K″, L″) Summary roses of the superimposed individual vectors for the indicated cells shown in panels K and L. Note the difference in appearance between mutant cells 2 and 3 and the other cells. EXPRESSION / LABELING:
PHENOTYPE:
|
|
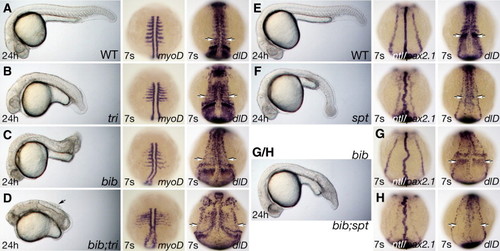
Interactions of bib with tri and spt. (A?D) Sibling embryos: 24 h phenotype, arrow indicates dorsal detached cells and 7-somite phenotype visualized with the myoD or dlD probes, arrows indicate lateral row of neurons. (E?H) Sibling embryos 24 h phenotype and 7-somite phenotype visualized with the ntl and pax2.1 or dlD probes, arrows indicate area of paraxial mesoderm expression. For bib and bib;spt in G/H, we could not distinguish the 24 h phenotypes. EXPRESSION / LABELING:
PHENOTYPE:
|
|
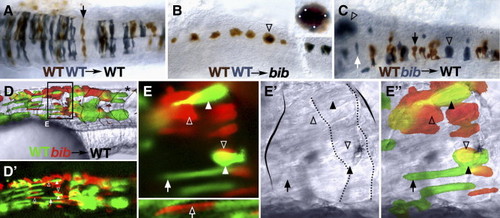
Autonomous and non-autonomous effects of bib in the nervous system and muscle. 24 h mosaic embryos after transplantation of wild-type and/or bib mutant cells into wild-type or bib mutant hosts as indicated. (A?C) Neural transplants into the hindbrain; dorsal view. (A) Arrow indicates typical wild-type neuroepithelial cell. (B) Arrow indicates aggregate of > 10 wild-type cells; inset shows magnification of cluster and white dots indicate individual cells. (C) Arrowheads indicate aggregate mutant cells, white arrow indicates isolated mutant cell, black arrow indicates normal wild-type cell. (D?E) Myotomal transplants into the trunk; lateral view. (D) Low magnification of chimera; dotted lines indicate myotome boundaries; asterisk indicates normal-shaped myotome. (E) Region magnified from panel D, showing UV, white light and composite images; solid arrow indicates typical wild-type fiber, hollow arrow indicates isolated normal-looking mutant fiber, solid arrowheads indicate abnormal wild-type fibers, hollow arrowheads indicate typical mutant aggregates; solid outline indicates abnormal myotome boundaries and dotted outline indicates unusual borders within the myotome. |
|
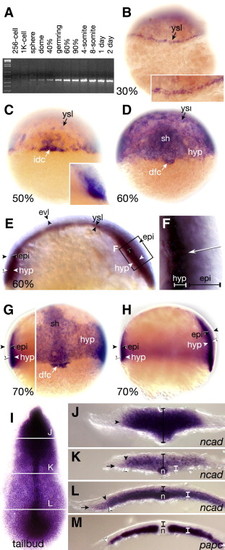
N-cadherin expression during epiboly and segmentation. (A) RT?PCR products. (B?L) In situ hybridizations with the N-cadherin probe. (B) Presumptive dorsal view; inset shows magnified view of yolk syncytial layer. (C) Dorsal view; inset shows magnified side view of deep cells. (D) Dorsal view. (E) Medial sagittal section, dorsal is right; arrowheads demarcate specific layers of the blastoderm. (F) Magnified view of boxed region in panel E, arrow indicates radial gradient of N-cadherin transcripts. (G) Dorsal view: left side, deep optical cross-section; right side, superficial view of same embryo. (H) Side view of panel G, deep optical cross-section, dorsal is right. (I) Flat mount dorsal view, lines indicate levels corresponding to transverse cross-sections J?L. (J) Head section, (K) trunk section, and (L) tailbud section. For panels J?L, black brackets demarcate neural plate, white brackets demarcate paraxial mesoderm, black arrowheads indicate edge of neural plate, white arrowheads indicate edge of paraxial mesoderm, and arrow indicates edge of lateral plate mesoderm. (M) Transverse cross section of a same stage embryo probed with papc. Abbreviations: dorsal forerunner cells, dfc; epiblast, epi; enveloping layer, evl; hypoblast, hyp; interior deep cells, idc; shield, sh; notochord, n; yolk syncytial layer, ysl. |
Reprinted from Developmental Biology, 310(2), Warga, R.M., and Kane, D.A., A role for N-cadherin in mesodermal morphogenesis during gastrulation, 211-225, Copyright (2007) with permission from Elsevier. Full text @ Dev. Biol.