- Title
-
whitesnake/sfpq is required for cell survival and neuronal development in the zebrafish
- Authors
- Lowery, L.A., Rubin, J., and Sive, H.
- Source
- Full text @ Dev. Dyn.
|
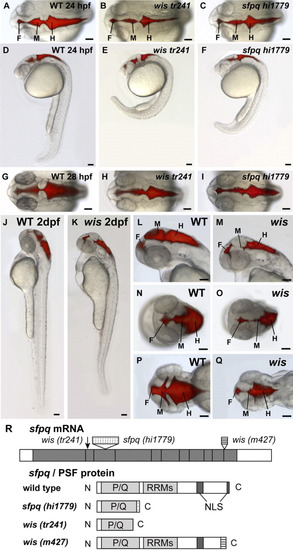
Phenotype of whitesnake/sfpq mutants. A-Q: Brain ventricles were visualized by microinjecting a fluorescent dye, Rhodamine-dextran, into the hindbrain ventricle of living anesthetized embryos at 24 hours postfertilization (hpf, A-F), 28 hpf (G-I), and 2 dpf (J-Q). A-F: At 24 hpf, the wistr241 mutant has reduced brain ventricles and abnormal curvature of the tail (B,E), as compared with wild-type (A,D), and the sfpqhi1779 mutant phenotype is similar (C,F). G-I: At 28 hpf, both wistr241 and sfpqhi1779 show variable reduction in brain ventricle size and reduced pigmentation. J-Q: By 48 hpf, the wis brain ventricle reduction becomes more severe compared with wild-type, especially in the midbrain. A-C,G-I,N-Q: Dorsal views. D-F,J-M: Side views. F, forebrain; M, midbrain; H, hindbrain. Scale bar = 100 μm. R:sfpq gene/PSF protein and corresponding mutations. wistr241 has a C to T mutation at position 491 of coding sequence, which results in an early stop codon at amino acid 167. sfpqhi1779 has a 6-kb retroviral insertion (which has a stop codon early in the insertion sequence). wism427 mRNA has aberrant splicing resulting in 200 base pairs of intronic sequence inserted before the last exon. Both sfpqhi1779 and wistr241 are truncated near the end of P/Q-rich region, and wism427 lacks the last NLS. P/Q, proline/glutamine-rich region; RRMs, RNA recognition motifs; NLS, nuclear localization sequence. PHENOTYPE:
|
|
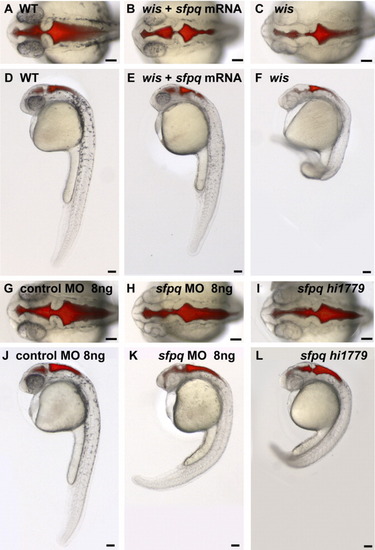
sfpq mRNA partially rescues the whitesnake phenotype and sfpq morpholino phenocopies mutant phenotype at 28 hours postfertilization (hpf). A-F: Wild-type (WT) sibling of wistr241 (A,D), wistr241 mutant injected with ∼100 pg sfpq mRNA (B,E), and wistr241 mutant (C,F). G-L: WT injected with 8 ng of control morpholino (G,J), WT injected with 8ng sfpq morpholino (H,K), and sfpqhi1779 mutant (I,L). A-C,G-I: Dorsal views. D-F,J-L: Lateral views. Scale bar = 100 μm. PHENOTYPE:
|
|
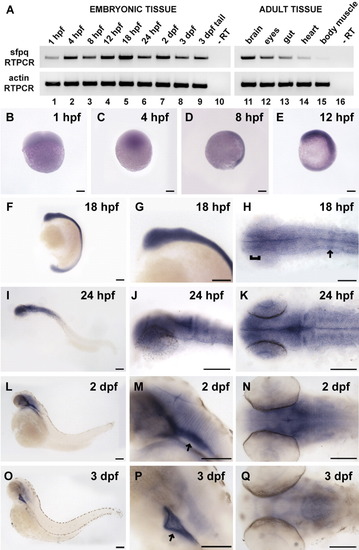
sfpq expression patterns. A: Reverse transcriptase-polymerase chain reaction (RT-PCR) for sfpq in embryonic tissue shows that sfpq is expressed beginning at 1 hpf (lane 1) and peaks at 18 hours postfertilization (hpf, lane 5). Adult tissue also has sfpq expression, with the brain showing the highest level (lane 11) and heart and body muscle showing lower levels (lanes 14, 15). Actin RT-PCR was used as control for RNA levels. B-Q: sfpq in situ hybridization time course shows that sfpq is expressed throughout embryogenesis. B: At 1 hpf (four-cell stage), side view. C: At 4 hpf (blastula stage), side view. D: At 8 hpf (75% epiboly stage), side view, dorsal right. E: At 12 hpf (six-somite stage), side view, dorsal right, anterior top. F-H: At 18 hpf, strongest expression in the forebrain (H, bracket) and in presumptive rhombomere 4 (H, arrow). I-K: At 24 hpf. L-N: At 2 dpf, expression appears restricted to strong longitudinal strips in the ventral brain (M, arrow) and in weaker transverse stripes in the hindbrain. O-Q: At 3 dpf, expression ventral to brain (P, arrow) and along axon tracts. F-G,I-J,L-M,O-P: Side views, anterior left. H,K,N,Q: Dorsal views, anterior left. Note: in I-Q, regions without staining may not be in focus, as to allow high magnification imaging of the stained regions. In areas of staining, the fuzziness sometimes observed is due to low levels of diffuse staining, not poor imaging. Scale bar = 100 μm. |
|
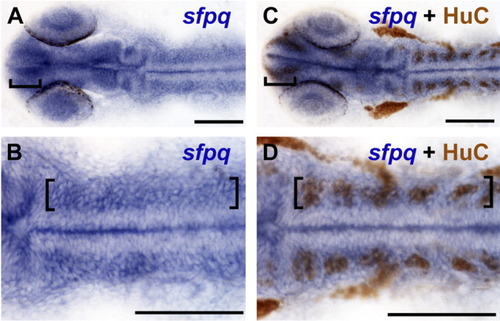
sfpq is strongly expressed in regions of neurogenic activity. A-D: At 24 hours postfertilization (hpf) wild-type embryos labeled for sfpq expression by in situ hybridization only (A,B) and wild-type sibling embryos double labeled for sfpq mRNA expression and HuC protein by immunohistochemistry (C,D) show that regions of strongest sfpq expression in the brain (brackets) overlap with HuC, a marker for postmitotic neuronal precursors. B,D: Higher magnification of hindbrain; brackets mark strong sfpq expression overlapping with HuC labeling. Midline staining is an artifact of the staining process, because it is not observed in embryos cut open before staining. A-D: Dorsal views. Anterior left. Scale bar = 100 μm. |
|
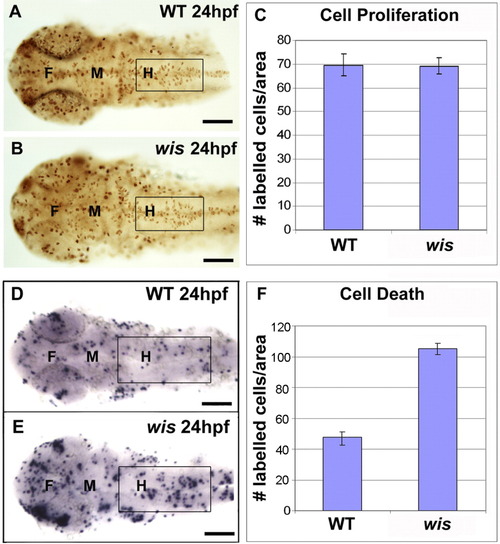
Cell proliferation and cell death analysis in whitesnake mutants. A-C: Cell proliferation analysis, using PH3 antibody labeling. A,B: Fixed and labeled wild-type and wis brain at 24 hours postfertilization (hpf). C: Quantification comparing labeling in hindbrain shows no difference between wild-type and mutant, n = 8; P = 0.8046. D-F: Cell death analysis, using terminal deoxynucleotidyl transferase-mediated deoxyuridinetriphosphate nick end-labeling (TUNEL) staining. D,E: Fixed and labeled wild-type and wis brain at 24 hpf. F: Quantification comparing labeling in hindbrain shows approximately twice the amount of cell death in the mutant than in wild-type, n = 14; P < 0.0001. Error bars denote standard error. A-B,D-E: Dorsal views. Boxes mark regions used for quantitation. F, forebrain; M, midbrain; H, hindbrain. Scale bar = 100 μm. PHENOTYPE:
|
|
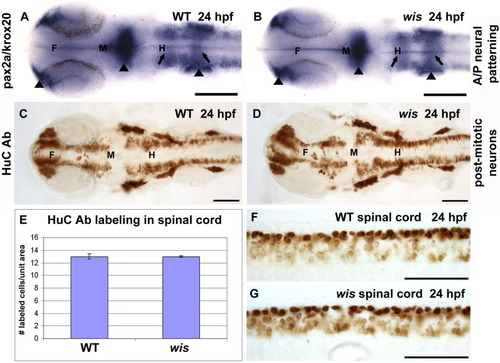
Neuronal determination is normal in whitesnake mutants. A,B: In situ hybridization for pax2a (labeling nasal placodes, midbrain-hindbrain boundary, and otic vesicles, arrowheads) and krox20 (labeling rhombomeres 3 and 5, arrows) show similar staining patterns in wild-type and mutant. C,D: Immunohistochemistry for HuC, a marker for postmitotic neurons, shows identical patterns between wild-type and mutant. E-G: This finding has been quantified in graph E, which depicts the number of HuC-labeled cells per unit area in the spinal cords of wild-type and wis (F,G). F, forebrain; M, midbrain; H, hindbrain. Scale bar = 100 μm. |
|
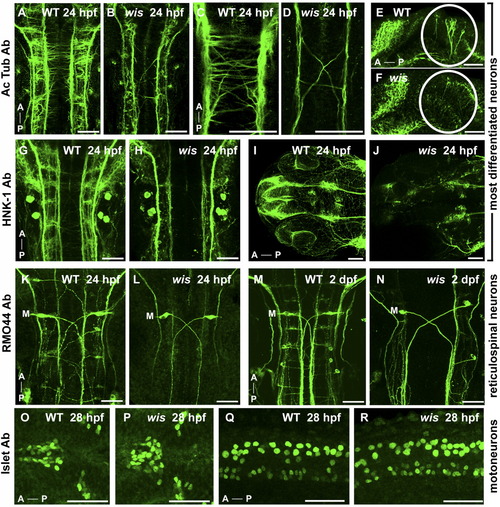
Specific neurons are absent in whitesnake mutants. A-J: Acetylated tubulin antibody labeling (A-F) and HNK-1 antibody labeling (G-J), both of which mark most differentiated neurons and their axons, show reduced number of axons in wis mutant in the hindbrain (B,D,H), midbrain (F), and eyes/forebrain (J) compared with wild-type (A,C,E,G,I). K-N: The RM044 Ab, which labels reticulospinal neurons, shows an absence of all reticulospinal neurons in the wis hindbrain except Mauthner neurons (labeled with M) at both 24 hours postfertilization (hpf, L) and 2 days postfertilization (dpf, N), compared with wild-type (K,M). O-R: The 4D5 Islet antibody, labeling motoneurons, shows no loss of motoneurons in whitesnake in the dorsal diencephalon (P) or spinal cord (R) compared with wild-type (O,Q). A, anterior; P, posterior. A-D,G-P: Dorsal views. E-F,Q-R: Side views. Scale bar = 50 μm. PHENOTYPE:
|