- Title
-
Large-scale enhancer detection in the zebrafish genome
- Authors
- Ellingsen, S., Laplante, M.A., Konig, M., Kikuta, H., Furmanek, T., Hoivik, E.A., and Becker, T.S.
- Source
- Full text @ Development
|
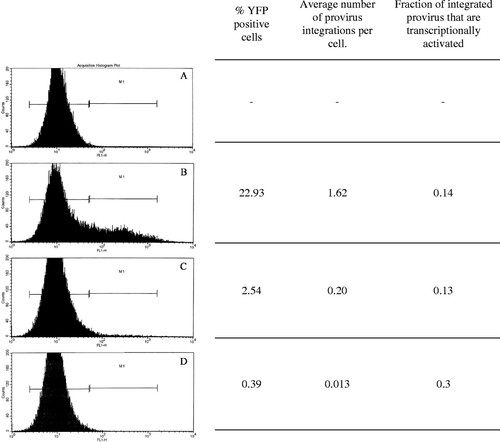
FACS analysis of zebrafish fibroblast cells (Pac2) infected with serial dilutions of the viral vector CLGY. Cells were infected with pseudotyped virus. Two days post infection, cells were harvested and analyzed for YFP expression by flow cytometry using a FACSCalibur Flow Cytometry Analyzer (Becton Dickinson, USA). (A) Uninfected cells. (B) Cells infected with concentrated CLGY stock (1.1x108 CFU/ml). (C) Cells infected with 1:10 dilution of concentrated virus stock. (D) Cells infected with 1:100 dilution of concentrated virus stock. To calculate average number of provirus integrations per cell, real-time quantitative PCR was performed on genomic DNA isolated from infected cells. Genomic DNA from cells containing two copies of an integrated provirus per cell was used as reference for provirus number, while variations in template amount were normalized against an endogenous target sequence (Chen et al., 2002) |
|
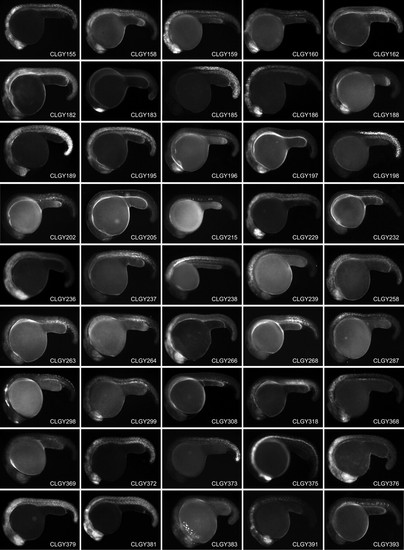
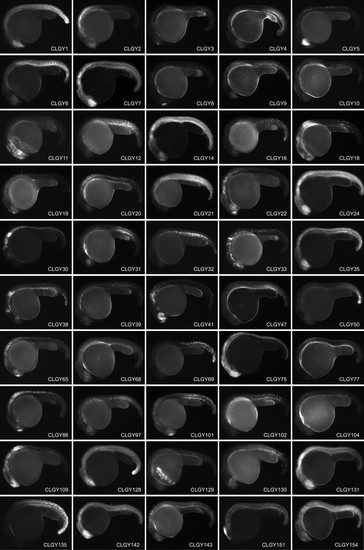
Stable transgenic lines showing spatially and temporally restricted expression of YFP around 24h post fertilization, anterior is to the left and dorsal to the top. Main domains of expression at 1 dpf (CLGY number is in parentheses): (1) Widespread expression, most pronounced in hypothalamus and posterior somites. (2) Diencephalon, hindbrain and anterior spinal cord. (3) Olfactory placodes. (4) Yolk syncytial layer, ventral caudal mesoderm. (5) Retina, epiphysis and spinal cord. (6) Developing skeletal muscle. (7) Widespread in CNS and muscle. (8) Dorsal telencephalon. (9) Hindbrain and posterior mesenchymal cells. (10) Subset of cells in CNS and notochord. (11) Neural crest and cranial ganglia. (12) Differentiating skeletal muscle and notochord. (14) Widespread, pronounced in telencephalon, retina, hindbrain and most posterior tissues. (16) Tail tip, notochord and skeletal muscle. (18) CNS, hindbrain (19) Dorsal telencephalon and mottled expression in notochord and spinal cord. (20) Hindbrain and spinal chord. (21) Developing skeletal muscle. (22) Telencephalon, olfactory placodes, hindbrain and spinal cord. (24) Widespread expression in CNS and muscle. (30) Hindbrain and spinal cord. (31) Retina and differentiating muscle. (32) Notochord and differentiating muscle. (33) Retina, hindbrain and spinal cord. (35) Widespread expression in CNS and muscle. (38) Retina, mid-hindbrain and hindbrain. (39) Sheath cells of the notochord (41) Retina, telencephalon and hindbrain. (47) Posterior mesenchymal cells. (50) Tail tip. (65) Telencephalon, olfactory placodes, hindbrain and spinal cord. (68) Widespread. (69) Spinal cord, posterior to somite 4/5 boundary. (75) CNS. (77) Spinal cord. (86) Telencephalon, hindbrain and spinal cord. (97) Notochord and differentiating muscle. (101) Telencephalon and differentiating muscle. (102) Differentiating skeletal muscle. (104) Cells in hindbrain and anterior part of spinal cord. (109) Forebrain and spinal cord. (128) Widespread, pronounced in ventral diencephalon and tailbud. (129) Hatching glands. (130) Retina and differentiating muscle. (131) Blood, retina, telencephalon, hindbrain, and muscle. (135) Posterior trunk. (142) Widespread. (143) Skeltal muscle and CNS. (151) Dorsal diencephalon and dorsal mesencephalon. (154) Widespread, strong in retina and telencephalon. (155) Telencephalon, retina, midbrain, rhombomeres, spinal cord. (158) Widespread. (159) Central nervous system and developing muscle. (160) Telencephalon and diencephalon. (182) Widespread in CNS and muscle. (183) Telencephalon. (185) Spinal cord and posterior trunk. (186) Central nervous system. (188) Retina and telencephalon. (189) Widespread. (195) Telencephalon, ventral diencephalon, MHB, hindbrain and spinal cord. (196) Ventral forebrain and retina. (197) Telencephalon and retina. (198) Spinal cord and posterior trunk. (202) Differentiating muscle. (205) Cells in forebrain, hindbrain and spinal cord. (215) Cells in notochord. (229) Widespread, strong in retina and hindbrain. (232) Mesenchymal cells (236) Retina and CNS. (237) Skeletal muscle. (238) Posterior hindbrain and spinal cord. (239) Hindbrain and spinal cord. (258) Widespread expression. (263) Retina and posterior mesenchyme. (264) Weak widespread. (266) Widespread, pronounced in CNS. (268) Differentiating muscle. (287) Hindbrain and spinal cord. (299) Retina, telencephalon and posterior mesenchymal cells. (308) Isolated cells in mesenchyme. (318) Telencephalic cells, neural crest and posterior mesenchyme. (368) Telencephalon, olfactory placodes, hindbrain and spinal cord. (369) Retina, notochord and differentiating muscle. (372) Blood, retina, telencephalon, hindbrain, and muscle. (373) Posterior trunk. (375) Ventral medial CNS. (376) CNS, strongest in retina, hindbrain and anterior spinal cord. (379) Widespread. (381) Widespread, strongest in CNS. (383) Retina and hatching glands. (391) Diencephalon and hindbrain. (393) Notochord. |
|
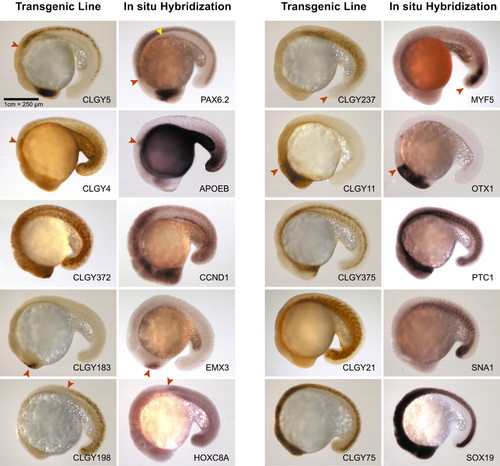
Comparison of YFP expression in transgenic lines with RNA expression patterns of candidate genes. Embryos are 18 hours post fertilization, anterior towards the left and dorsal upwards. CLGY5/pax6.2 are expressed in the same domains, retina, pineal and spinal cord, with exception of the pancreas seen in the in situ stain (yellow arrowhead) and much weaker expression in hindbrain in the transgenic line (anterior limit of expression domain is marked with a red arrowhead). CLGY4/apoeb are highly similar in expression, including the pectoral fin buds (arrowhead). CLGY372/ccnd1 are very similar with a widespread and highly dynamic expression. CLGY183/emx3 are expressed in the telencephalon only (arrowhead). CLGY198/hoxc8a share the same anterior boundary (arrowhead). Expression in CLGY237/myf5 is similar in the developing muscle, but the tail bud expression (arrowhead) of the endogenous RNA is not seen in the transgenic line. CLGY11/otx1l have a highly similar expression pattern, anterior boundary shown by an arrowhead. CLGY375/ptc1 are very similar. CLGY21/snai1a are expressed in the same domain, skeletal muscle. CLGY75/sox19 are both expressed at high levels throughout the CNS. |
|
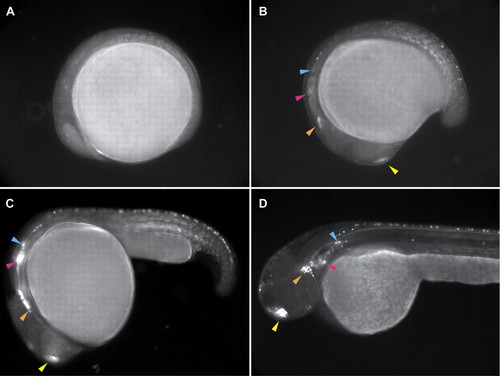
Expression in sensory placodes in CLGY298. (A) 8 hpf; (B) 16 hpf; (C) 24 hpf; (D) 48 hpf. Anterior is towards the left. Expression of YFP is first seen at 16 hpf in the olfactory placode (yellow arrowhead), the trigeminal placode (orange arrowhead), the inner ear (pink arrowhead) and the lateral line primordium (blue arrowhead). At 24 hours the latter three primordia migrate together before reaching their final destination, as seen at 48 hours (D). Expression is also seen in skeletal muscle early on, as well as in sensory neurons in the spinal cord. |