- Title
-
Transcriptional regulation of mitfa accounts for the sox10 requirement in zebrafish melanophore development
- Authors
- Elworthy, S., Lister, J.A., Carney, T.J., Raible, D.W., and Kelsh, R.N.
- Source
- Full text @ Development
|
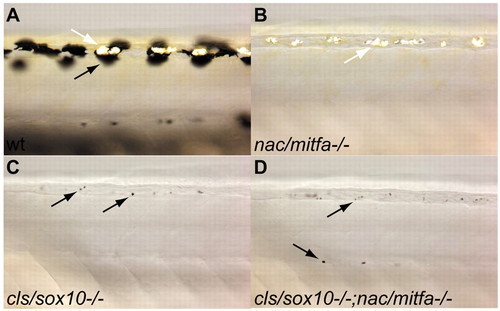
Pigment cell defects in cls/sox10-/-, nac/mitfa-/- and cls/sox10-/-;nac/mitfa-/- mutant embryos. Lateral views of the dorsal trunk of 3 dpf wild type (A), nac/mitfab692/b692 (B), cls/sox10t3/t3 (C) and cls/sox10 t3/t3;nac/mitfa b692/b692 (D) embryos. Wild-type embryos have large flat melanophores (black arrow), cls/sox10-/- and cls/sox10-/-;nac/mitfa-/- embryos have a few tiny rounded melanophores (black arrows), but nac/mitfa-/- embryos lack melanophores. Iridophores (white arrows) are not reduced in nac/mitfa-/- embryos but are severely reduced in cls/sox10-/- and cls/sox10-/-;nac/mitfa-/-. Double mutant embryos were identified by PCR genotyping. PHENOTYPE:
|
|
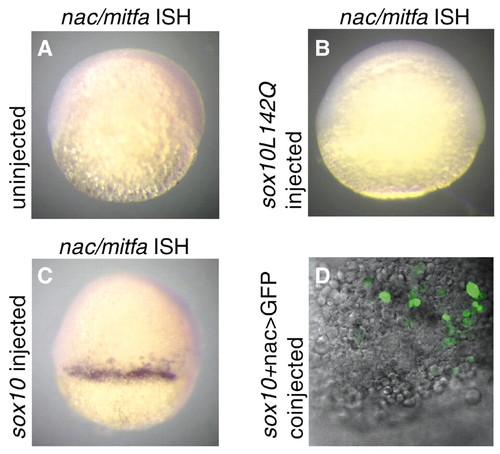
Precocious nac/mitfa expression in 6 h.p.f embryos following injection with cls/sox10 RNA. Lateral views of uninjected (A), cls/sox10L142Q RNA injected (250 pg per embryo; B) and cls/sox10 RNA injected (250 pg per embryo; C) 6 hpf embryos following in situ hybridization with a nac/mitfa probe. Spots and/or patches of nac/mitfa expression were detected in 39% of cls/sox10 RNA injected embryos (n=136) but not in any of the uninjected embryos (n=58) nor in any of the cls/sox10L142Q RNA injected embryos (n=92). (D) Superimposed fluorescent confocal and DIC images of an animal/lateral view of a 6 hpf embryo coinjected with cls/sox10 RNA (250 pg per embryo) and nac>GFP reporter plasmid (150 pg per embryo) show cells with GFP fluorescence. GFP fluorescence was observed in 75% (n=224) of embryos coinjected with cls/sox10 RNA and nac>GFP. |
|
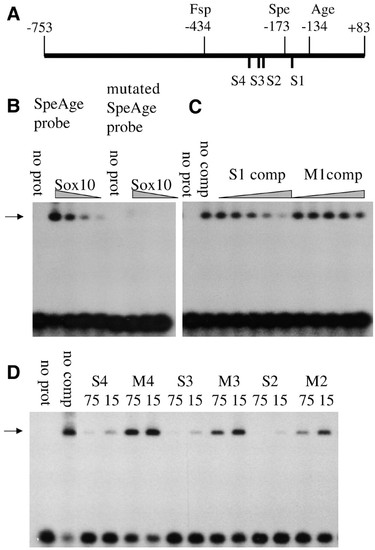
Electrophoretic mobility shift assays (EMSA) showing binding of Cls/Sox10-GST fusion protein to sites in the nac/mitfa promoter. (A) Schematic diagram of the 836 b.p. nac/mitfa promoter showing the positions of the putative sox binding sites S1, S2, S3 and S4. (B) The Spe1-Age1 fragment of the nac/mitfa promoter (SpeAge probe) shows a band of reduced electrophoretic mobility (black arrow) with ∼20 nM, 10 nM, 5 nM and 2.5 nM Cls/Sox10-GST fusion protein (Sox10) which is not seen without the Cls/Sox10-GST protein (no prot). When site S1 is mutated in this DNA fragment (mutated SpeAge probe) binding under these same Cls/Sox10-GST protein concentrations is greatly reduced. (C) Binding of∼ 10 nM Cls/Sox10-GST protein to the Spe1-Age1 fragment of the nac/mitfa promoter is effectively competed by an oligonucleotide with site S1 (S1 comp) but not by the mutated site oligonucleotide M1 (M1 comp). Shown are binding reactions with a serial five-fold dilution series of this competitor oligonucleotide giving 0.13 to 75 pmoles per reaction and also controls with no specific competitor (no comp) and with no Cls/Sox10-GST protein (no prot). (D) Binding of ∼10 nM Cls/Sox10-GST protein to the Spe1-Age1 fragment of the nac/mitfa promoter is effectively competed by oligonucleotides with binding sites S2, S3 or S4 but less effectively by the mutated versions M2, M3 or M4. Shown are binding reactions with 75 pmoles (75) or 15 pmoles (15) of these competitor oligonucleotides and also controls with no specific competitor (no comp) and with no Cls/Sox10-GST protein (no prot). |
|
In vivo melanophore rescue by forced nac/mitfa expression in the premigratory neural crest. Lateral views of the posterior trunk of 2 dpf, cls/sox10-/- (A) and nac/mitfa-/- (B) embryos that had been injected with cls>nac (15 pg per embryo). Rescued melanophores (arrows) have normal position and morphology. |