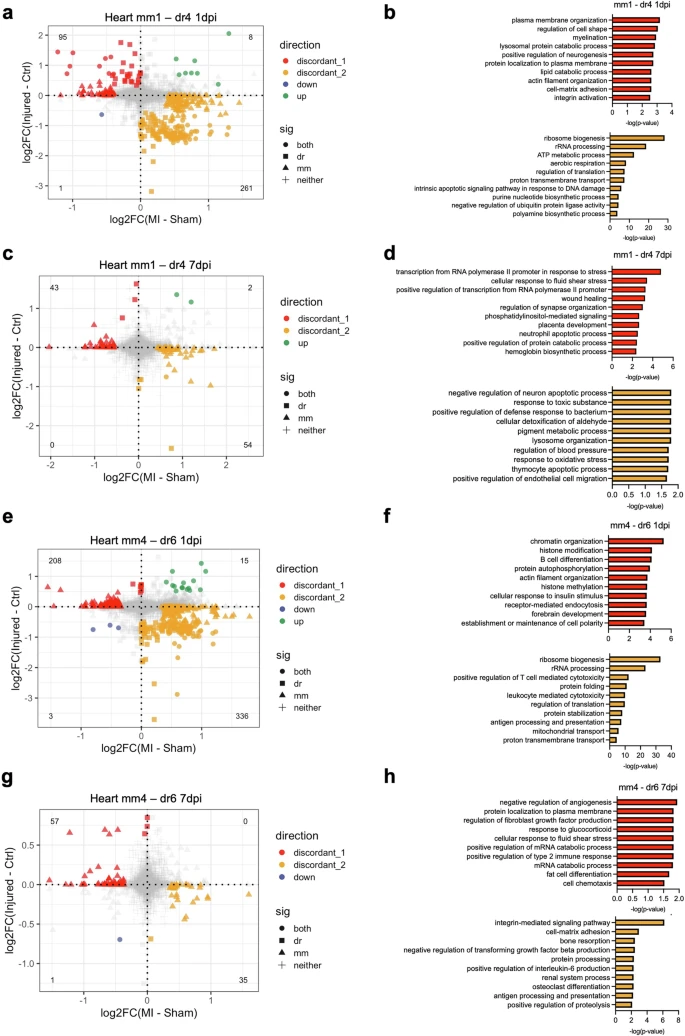
Fig. 5 Differential transcriptional response to cardiac injury in mono/M? subsets between species.a Scatterplot of the fold changes in murine and zebrafish genes with cardiac injury compared to control in mm1 and dr4 cells at 1 dpi (FDR < 0.10 and |log2FC| > 0.25). b Enriched biological process terms of the differentially expressed genes (DEGs) that are discordant between mm1 and dr4 cells in (a). c Scatterplot of the fold changes in murine and zebrafish genes with cardiac injury compared to control in mm1 and dr4 cells at 7 dpi (FDR < 0.10 and |log2FC| > 0.25). d Enriched biological process terms of the DEGs that are discordant between mm1 and dr4 cells in (c). e Scatterplot of the fold changes in murine and zebrafish genes with cardiac injury compared to control in mm4 and dr6 cells at 1 dpi (FDR < 0.10 and |log2FC| >0.25). f Enriched biological process terms of the DEGs that are discordant between mm1 and dr4 cells in (e). g Scatterplot of the fold changes in murine and zebrafish genes with cardiac injury compared to control in mm4 and dr6 cells at 7 dpi (FDR < 0.10 and |log2FC| >0.25). h Enriched biological process terms of the DEGs discordant between mm1 and dr4 cells in g. In (a), (c), (e), and (g), concordant genes with FDR < 0.10 and |log2FC| > 0.25 in both species were colored blue when downregulated and green when upregulated. Discordant genes with FDR < 0.10 and |log2FC| > 0.25 in one specie were colored red when upregulated in zebrafish and yellow when upregulated in mouse. In (b), (d), (f), and (h), pathways with a -log10(p-value) >1.3 were considered significantly enriched.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Commun Biol