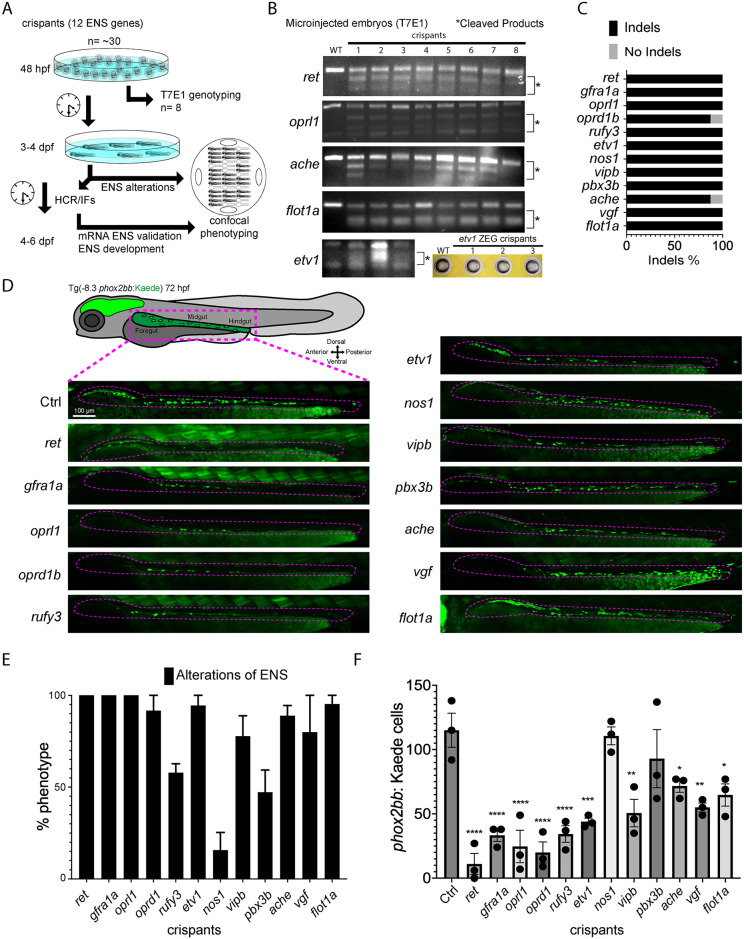
Fig 2 Candidate genes targeted in an F0 CRISPR screen display ENS phenotypic alterations.
(A) At 48 hpf, eight crispant embryos from pools of around thirty embryos were used to validate CRISPR activity via T7E1, for each gene targeted. If the majority had indels, then subsets of the pool were grown at 3 to 4 dpf to phenotype their ENS. The phenotyping process combined crispants of different genes by using an agarose cast that enabled high-content semi-automated confocal imaging. An additional fraction of the crispants were analyzed at 4–6 dpf for additional HCR validation or for late phenotypic alterations. (B) Representative images of different T7E1 assays demonstrate indels of different embryos in the specific gene-targeted regions.