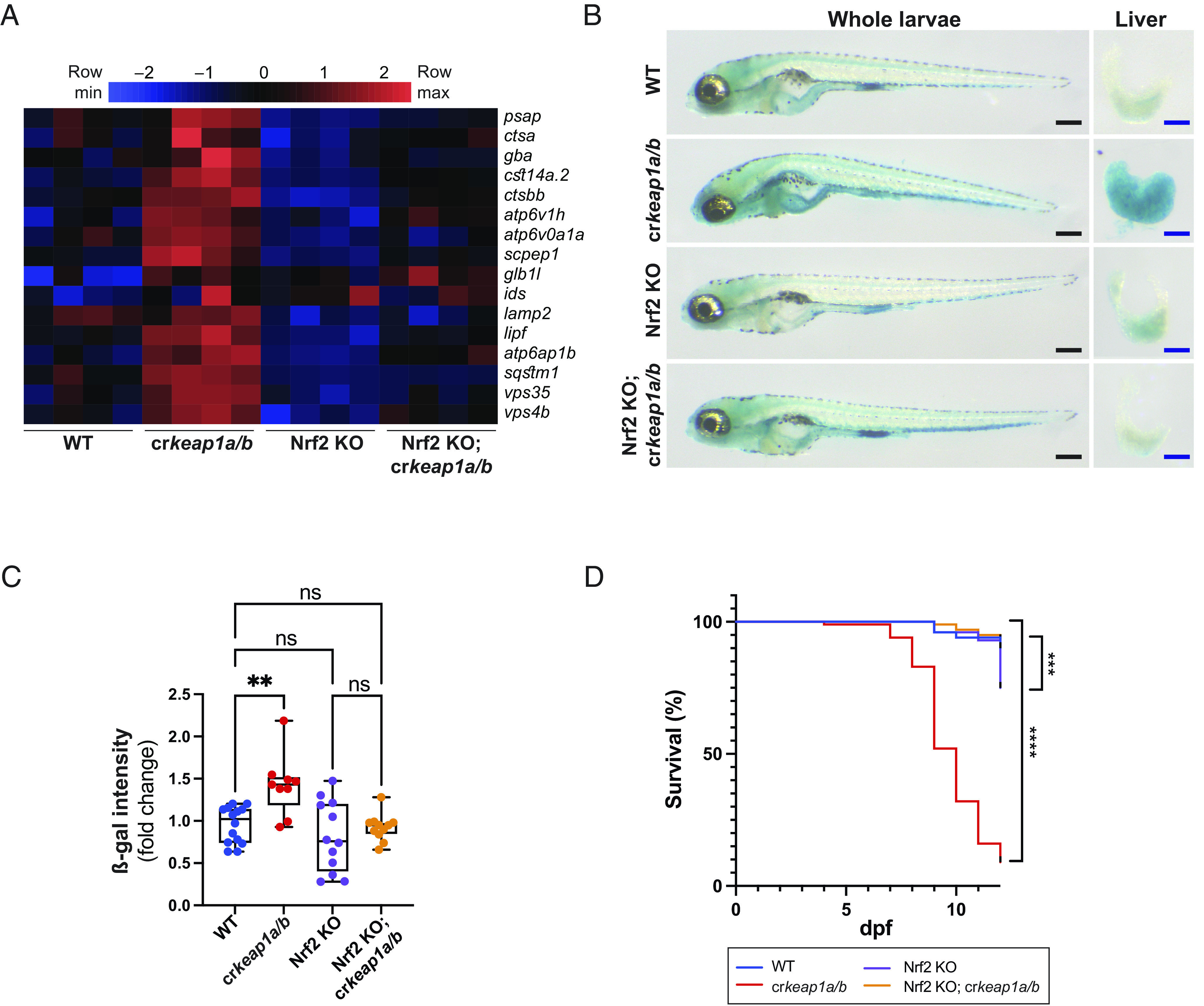
Fig. 5.
Keap1-dependent regulation of lysosomal biogenesis requires Nrf2. (A) Heatmap of lysosomal gene expression in WT, crkeap1a/b, Nrf2 KO, and Nrf2 KO; crkeap1a/b zebrafish at 7 dpf as determined by RNA-Seq analysis, n = 4 pools of 10 larvae. (B) Representative images of ß-gal-stained whole-mount larvae (Left) and dissected larval livers (Right) from WT, crkeap1a/b, Nrf2 KO, and Nrf2 KO; crkeap1a/b zebrafish at 7 dpf. Black scale bars represent 350 µm, blue scale bars represent 100 µm. (C) Quantification of ß-gal intensity in dissected livers in B. Data are shown as mean and interquartile range, n = 9 to 14 livers. (D) Kaplan–Meier survival plot of WT, crkeap1a/b, Nrf2 KO, and Nrf2 KO; crkeap1a/b zebrafish, n = 40. For all experiments, **P < 0.01, ***P < 0.001, ****P < 0.0001.