Fig. 7
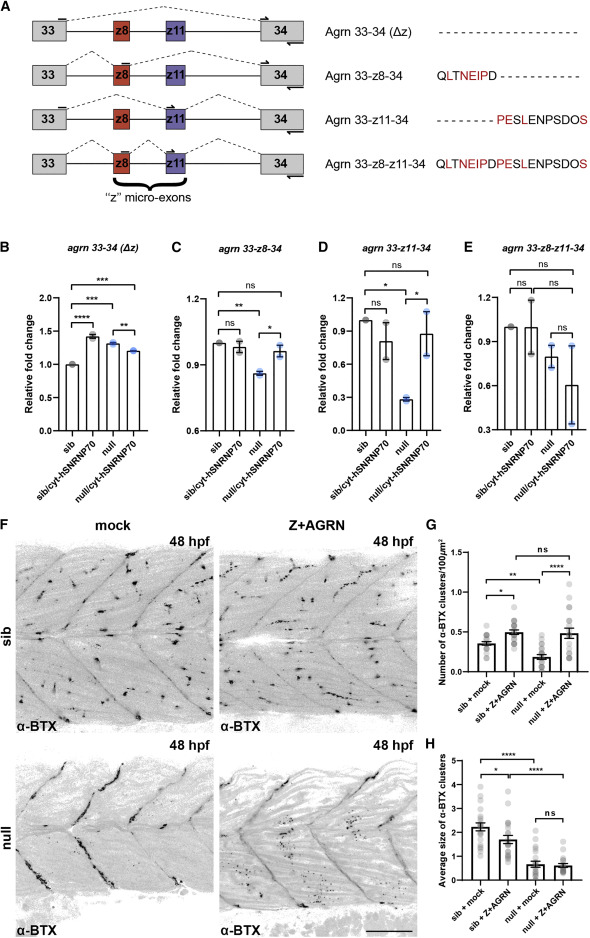
Figure 7. Cytoplasmic SNRNP70 regulates the abundance of Z+AGRN splice variants (A) Agrin Z+ (Z+AGRN) isoforms identified by RT-PCR are shown schematically, with conserved residues between humans and zebrafish shown in red. Arrows depict the position of primers used for the qPCR analysis in (B)?(E). (B?E) RT-qPCR analysis in sibling and null animals as well as following ubiquitous overexpression of cytosolic hSNRNP70-eGFP using Tg(ubi:ERT2-Gal4; UAS:hSNRNP70?NLS3xNES-eGFP) embryos at 28 hpf. Primers were designed to specifically amplify the expression of non-z+agrn (agrn 33?34 (?z)) isoforms (B), as well as the three z+agrn isoforms: 33-z8-34 (C), 33-z11-34 (D), and 33-z8-z11-34 (E). The graphs show mean values ± SEM for fold change in expression in relation to control animals (sib). ????p < 0.0001; ???p < 0.001; ??p < 0.01; ?p < 0.05; ns, not significant, one-way ANOVA, datapoints represent two independent biological replicates per group. (F) Representative images depicting a superficial layer of the trunk myotomes showing the distribution of ?-BTX-labeled AChR clusters in mock- and Z+AGRN-treated sibling and null animals at 48 hpf. Images are lateral views with anterior to the left. Scale bars, 50 ?m. (G) Quantification of the number of ?-BTX clusters in the four groups. The graph shows mean values ± SEM. ????p < 0.0001; ??p < 0.01; ?p < 0.05; ns, not significant, one-way ANOVA, n = 21 animals per group in two independent experiments. (H) Graph showing mean values ± SEM of the average size of ?-BTX clusters in the four groups. ????p < 0.0001; ?p < 0.05; ns, not significant, one-way ANOVA, n = 21 animals per group in two independent experiments.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Curr. Biol.