Figure 1
Figure 1
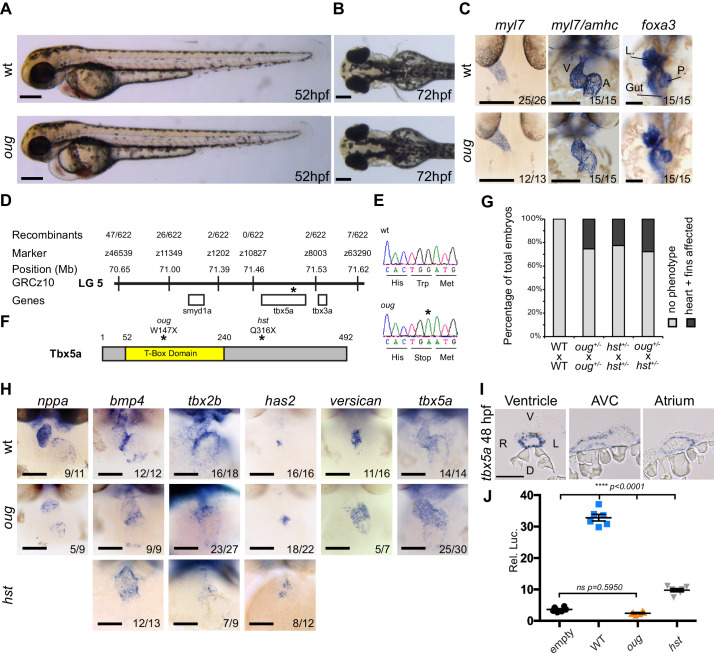
(A) Lateral view of wt and oug mutant embryos at 52 hpf. Note the cardiac edema in oug. (B) At 72 hpf dorsal observation of oug mutant embryos reveals absence of lateral fins. (C) Two dpf oug mutant embryos display defective cardiac looping but normal asymmetric positioning of the internal organs. L, liver; P, pancreas (D) Mapping and genomic position of the oug mutation (indicated by the asterix). (E) A single-nucleotide substitution in tbx5a (G to A) resulting in a tryptophan (Trp; TGG) to stop (TGA) mutation segregates with the oug phenotype. (F) Tbx5a is truncated at amino acid 147 in oug, in its T-Box domain. The hst allele (Q316X; Garrity et al., 2002) is included for comparison (G) Complementation test. Outcross of oug+/- to hst+/- fails to complement the oug cardiac and pectoral fin bud phenotype. (H) Gene patterning is affected in oug hearts at 2dpf. Expression of nppa is reduced in the cardiac chambers while expression of bmp4 and tbx2b is expanded in the AV canal. Cardiac cushion markers has2 and versican also show expanded expression domains. ISH for hst is shown for comparison: while bmp4 and has2 display expanded expression domains as in oug, tbx2b is barely detectable. Transcripts for tbx5a are detected in wt and oug mutants. (I) Transcripts for tbx5a can be detected evenly in transversal sections through the entire 2 dpf heart tube. (J) Luciferase assay establishes that oug retains virtually no activity. Mean values ▒ SEM are shown. Scale bars (A,B,C,H): 100 Ám; (I): 50 Ám.
The oudegracht (oug) mutant carries a tbx5a null allele and displays defective cardiac looping.