Fig. S1
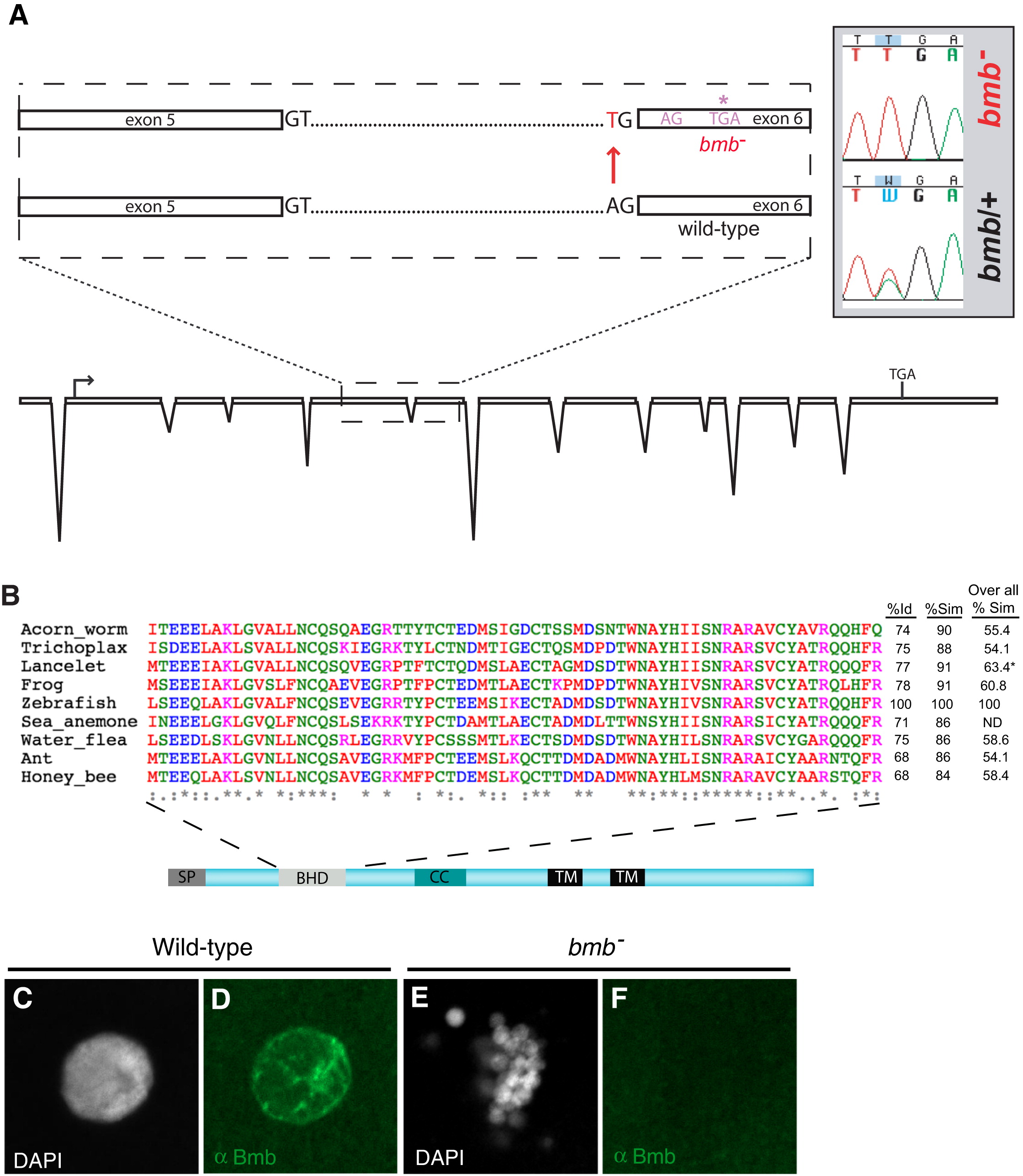
Cloning, Alignment of the BHD from Potential Homologs, and Anti-Bmb Staining of Interphase Nuclei, Related to Figure 3(A) Our annotation of non-gene encoding genomic regions in the interval revealed a gene corresponding to a novel ORF, ORF1.8. In this gene in the p22atuz mutant allele, we identified an A to T base change at the splice acceptor site upstream of exon 6.(B) A 69-residue domain spanning residues 91 to 159 of zebrafish Bmb is compared with a diverse set of potential homologs (first column- % identity (Id), second column- % similarity (Sim) with in the BHD. The third column is a comparison of overall similarity among the potential Bmb homologs. * Only the available residues of the lancelet protein are aligned. A significant portion of the sea anemone protein is not available so overall similarity was not determined (ND).(C?F) (C and D) WT and bmb (E and F) interphase selected embryos were stained with Dapi (C and E) and anti-Bmb (D and F). Embryos are at the 32-cell stage (n = 3 WT; n = 6 bmb). Images correspond to a Z-projection from a confocal Z-series.
Reprinted from Cell, 150(3), Abrams, E.W., Zhang, H., Marlow, F.L., Kapp, L., Lu, S., and Mullins, M.C., Dynamic Assembly of Brambleberry Mediates Nuclear Envelope Fusion during Early Development, 521-532, Copyright (2012) with permission from Elsevier. Full text @ Cell