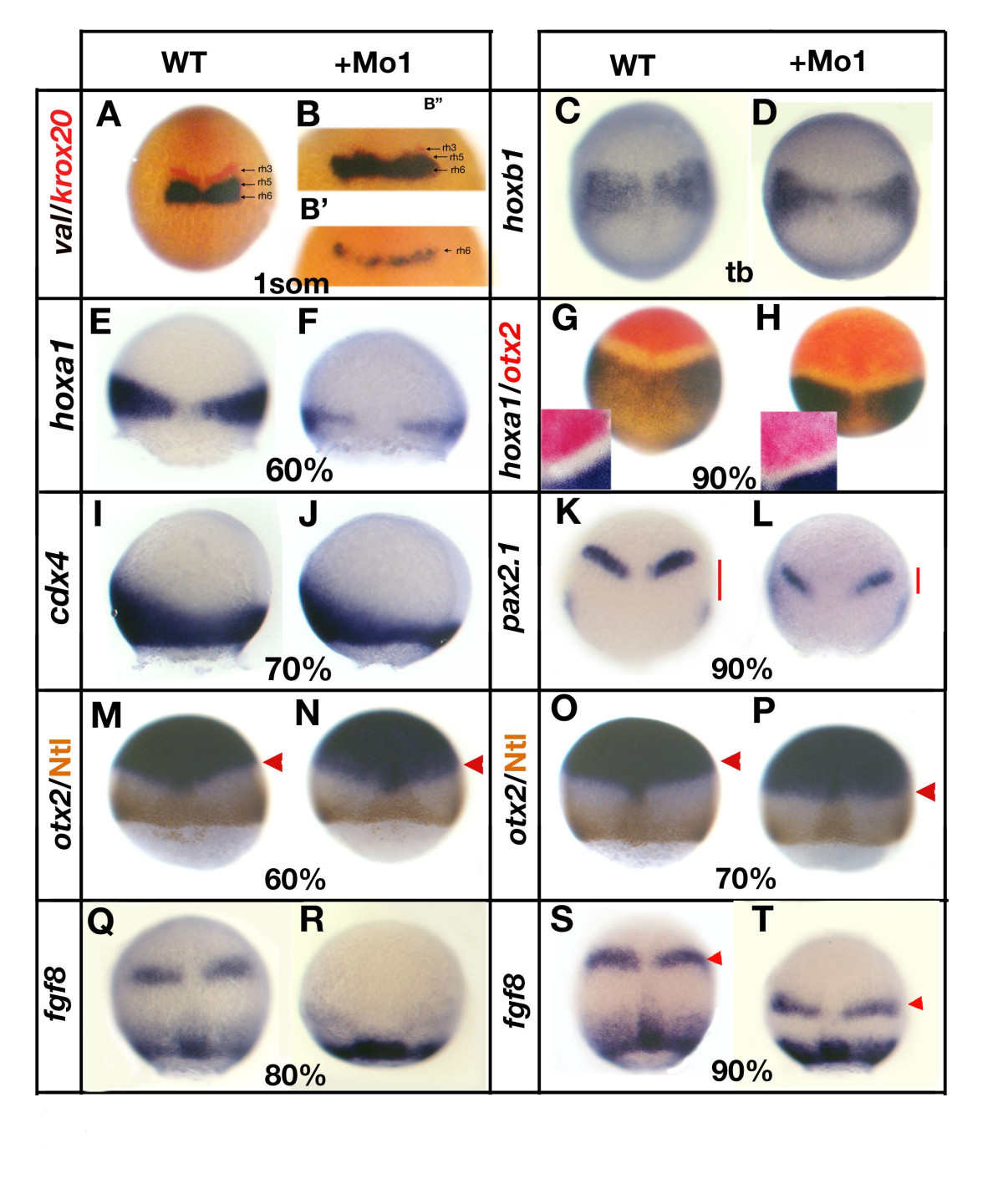
Fig. 6
Fig. 6 Loss of gbx1 affects hindbrain and midbrain-hindbrain boundary (MHB) markers. (A-H, M-K-T) are dorsal views, (I, J) are lateral views and the developmental stages are indicated (tb, tailbud stage). (A) Control embryo (wild type (WT)) stained with val (blue) and krox20 (red), (B, B′) gbx1-morphants. A residual krox20 expression is seen in (B′) and no expression of krox20 in (B″). For (B′, B″) a higher magnification of the region of interest is shown. (C) Control embryo stained with hoxb1; (D) gbx1-morphant. (E) Control embryo stained with hoxa1; (F) gbx1-morphant. (G) Control embryo stained with hoxa1 and otx2; (H) gbx1-morphant. A higher magnification is shown, illustrating the reduction of size of the gap between the otx2 and hoxa1 domains. (I) Control embryo stained with cdx4; (J) gbx1 morphant. (K) Control embryo stained with pax2.1; (L) gbx1-morphant. The red bars show the distance between the MHB and the lateral plate mesoderm expression domains. This distance is markedly reduced in the gbx1 morphants. (M, O) Control embryos stained with otx2 (blue) and Ntl antibody (brown); (N, P) gbx1 morphants. The otx2 domain is unaffected at 60% epiboly but slightly shifted posteriorly at 70% (red arrowheads). (Q, S) Control embryos stained with fgf8; (R, T) gbx1-morphants. MHB expression is delayed (R) and shifted posteriorly (S, T, red arrowheads) in the morphants.