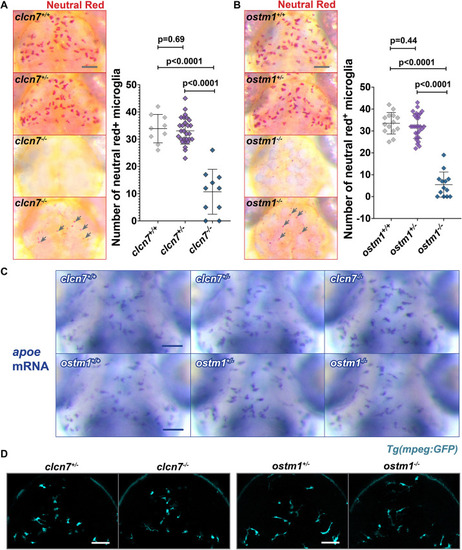
Reduced Neutral Red staining in clcn7 and ostm1 mutants. (A,B) Neutral Red labeling revealed significantly fewer microglia labeled with this vital dye in (A) clcn7 and (B) ostm1 mutants. Some mutants displayed small Neutral Red punctae (gray arrows). These smaller Neutral Red punctae were included in the quantification. clcn7+/+: n=9, mean=33.9, s.d.=5.2, s.e.m.=1.7. clcn7+/−: n=28, mean=33.1, s.d.=4.9, s.e.m.=1.0. clcn7−/−: n=10, mean=10.7, s.d.=8.2, s.e.m.=2.6. ostm1+/+: n=14, mean=33.5, s.d.=4.9, s.e.m.=1.3. ostm1+/−: n=28, mean=32.2, s.d.=5.6, s.e.m.=1.1. ostm1−/−: n=13, mean=5.5, s.d.=5.8, s.e.m.=1.6. Two-tailed unpaired t-test with Welch's correction was performed to calculate significance; graphs show mean±s.d. (C) apoe in situ hybridization showed that clcn7 and ostm1 mutants have normal numbers and morphology of microglia. (D) Confocal microscopy confirmed that mpeg1:GFP expression is normal in clcn7 and ostm1 mutants. Images in A, B and D are representative of at least three independent experiments, and those in C of two independent experiments. Scale bars: 50 µm.
|

