Fig. 1
- ID
- ZDB-FIG-230323-13
- Publication
- Alvizi et al., 2022 - mir152 hypomethylation as a mechanism for non-syndromic cleft lip and palate
- Other Figures
- All Figure Page
- Back to All Figure Page
|
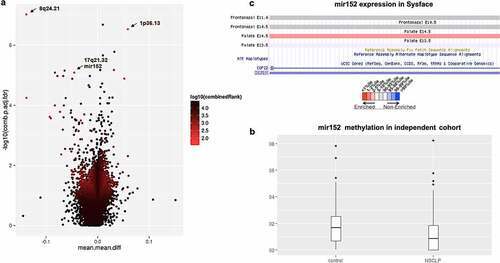
Figure 1. mir152 is differentially methylated in NSCLP cohorts. (a) Volcano plot of differentially methylated regions (DMRs) in the 450 K cohort. Light blue spots are the best ranked DMRs by a p-value, methylation difference and quotient of difference by RnBeads. Arrows indicate DMRs at 8q24.21, 1p36.13, and mir152. (b) mir152 is significantly hypomethylated in the BSAS cohort. Boxplots with central lines as medians. p-Value = 0.005 (Mann?Whitney?s test). (c) mir152 expression changes are detected in developing craniofacial structures as revealed at the database Sysface. Higher signals for mir152 expression are detected in developing murine palate (E14.5) in red and also frontonasal prominences E10.5 and E11 (grey). |