Fig. 1
- ID
- ZDB-FIG-221229-25
- Publication
- Klatt Shaw et al., 2021 - Efficient CRISPR/Cas9 mutagenesis for neurobehavioral screening in adult zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
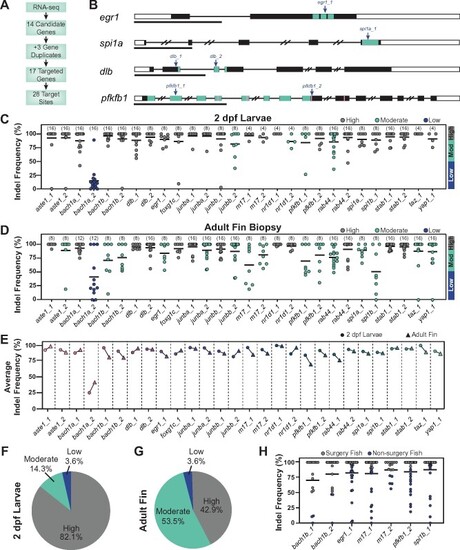
CRISPR/Cas9 dgRNPs effectively mutate genes in adult zebrafish. (A) Schematic summary of gene targeting. A total of 17 genes were targeted using 28 dgRNPs. (B) Representative schematics for dgRNP target genes. Shown are transcription factors egr1 and spi1a, the dlb ligand, and the enzyme pfkfb1 (black boxes, exons; white boxes, UTRs; and lines, introns). Key domains are indicated in teal: zinc finger (egr1) and ETS (spi1a) DNA binding domains, EGF-like domain 1 (dlb), and 6PF2K domain (pfkfb1). The enzymatic active sites are indicated by pink lines in pfkfb1. dgRNP target sites are indicated by blue arrows. Scale bars, 1 Kilobase (Kb). (C, D) Targeting efficiency in dgRNP injected zebrafish crispants by capillary electrophoresis. Data points represent individual animals . Lines indicate means. Sample sizes are indicated between parentheses. 2 dpf larvae (C) or adult caudal fin (D) were analyzed. Target sites are classified as high-efficiency (indel frequency >90%, gray), moderate-efficiency (indel frequency 50–90%, teal), and low-efficiency (indel frequency <50%, blue). (E) Targeting efficiency in larval (circles) and adult (triangles) zebrafish. Data points represent the average indel frequency for each target site. (F, G) Pie chart representation of dgRNP efficiency. Shown are the fractions of dgRNPs with high (gray), moderate (teal), and low (blue) efficiencies. (H) Capillary electrophoresis genotyping of adult animals in which indel frequency decreased between larvae and adults. Data points represent individual animals. Adult fish with indel frequency >90% (gray) were subjected to spinal cord transection and subsequent phenotyping. Because only 16 taz_1/yap_1 dual-targeted animals survived to adulthood, all were genotyped in D. |