Fig. 3
- ID
- ZDB-FIG-220906-3
- Publication
- Colijn et al., 2022 - High-throughput methodology to identify CRISPR-generated Danio rerio mutants using fragment analysis with unmodified PCR products
- Other Figures
- All Figure Page
- Back to All Figure Page
|
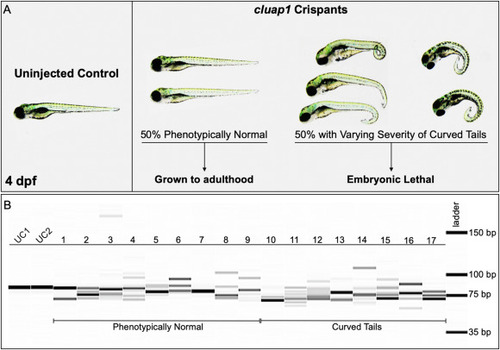
Fig. 3. cluap1 crispants display varying body curvature severity and successful CRISPR targeting. (A) Images of uninjected control (left) and cluap1 crispant embryos (right) at 4 dpf. Approximately 50% of the crispants developed the tail curvature and embryonic lethality indicative of homozygous mutations in cilia proteins (Cao et al., 2010; Li and Sun, 2011; Sullivan-Brown et al., 2008), though the severity of the tail curvature varied. The remaining phenotypically normal survivors were grown to adulthood (F0 generation) to use for breeding stable mutant lines. (B) Crispants were collected at 4 dpf and genotyped for the presence of indels with the fragment analyzer workflow. Image of the fragment analyzer gel output data is shown. The first two lanes (UC1, UC2) are uninjected sibling controls. Lanes 1?9 are crispants that appear phenotypically normal. Lanes 10?17 are crispants that display tail curvature. Only 1 out of 35 genotyped crispants displayed a single wildtype band (Lane 7), indicating a success rate of 97% (34/35) for indel generation. |
Reprinted from Developmental Biology, 484, Colijn, S., Yin, Y., Stratman, A.N., High-throughput methodology to identify CRISPR-generated Danio rerio mutants using fragment analysis with unmodified PCR products, 22-29, Copyright (2022) with permission from Elsevier. Full text @ Dev. Biol.