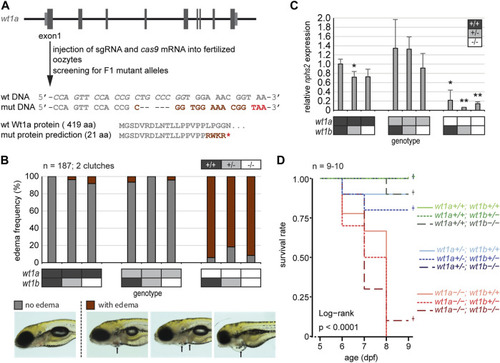
Inactivation of wt1a results in edema formation and early lethality. (A) Schematic drawing of the wt1a gene locus and mutant generation. With the help of CRISPR/Cas, a fish line was established that lacks five nucleotides in exon 1 of the wt1a gene (wt1a ex1_del5 ). The sgRNA target sequence is italicized in the wild type DNA sequence. The 5 nt deletion is predicted to result in a frame shift (dark red) and a premature termination codon (red) that leads to a truncation of the protein. Abbreviations: wt (wild type); mut (mutant). (B) Offspring of a double heterozygous incross (wt1a +/-;wt1b +/-) were screened for the formation of edema at five dpf. The majority of wt1a ?/? larvae have body edemas (arrows), independent of the wt1b ex2_del5 background. Example pictures for no edema (grey), and body edemas (dark red) are shown; n = 187. (C) qRT-PCR analysis of offspring of a double heterozygous incross (wt1a +/-;wt1b +/-) indicates a strong reduction of nphs2 expression in wt1a ?/? larvae. n = 3 (three to five larvae each). Error bars represent standard error. One-way ANOVA was calculated as p = 3.98*10?05. p-values are calculated with a t-test by pairwise comparison to wild type (wt1a +/+; wt1b +/+). (D) Survival of offspring of a double heterozygous incross (wt1a +/-;wt1b +/-) from five to nine dpf shows an early onset of lethality of homozygous wt1a ?/? larvae, independent of the wt1b ex2_del5 background. Survival is shown as a Kaplan-Meier curve. n = 9?10 larvae per genotype.