Figure 3
- ID
- ZDB-FIG-210915-78
- Publication
- de Vrieze et al., 2021 - Efficient Generation of Knock-In Zebrafish Models for Inherited Disorders Using CRISPR-Cas9 Ribonucleoprotein Complexes
- Other Figures
- All Figure Page
- Back to All Figure Page
|
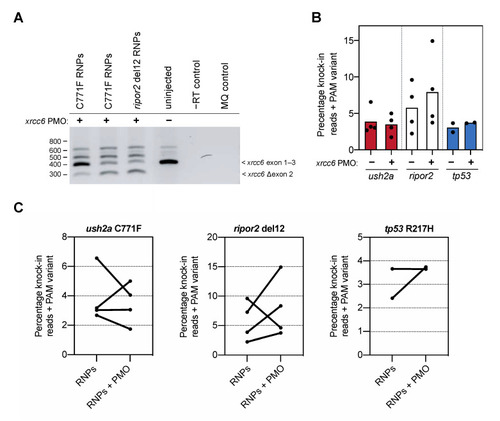
Knock-in efficiency after co-delivery of the xrcc6-targeting PMO. (A) Addition of the xrcc6-targeting PMO to the RNP injection mixtures results in alterative xrcc6 splicing. Both intron retention and exon 2-skipping are observed upon RT-PCR analysis of the xrcc6 transcript in PMO-injected embryos, but not control injected embryos. Replicate injections of C711F RNPs provide insight in variability of PMO-induced alterative xrcc6 splicing. (B) Quantification of knock-in efficiency after sgRNA-Cas9 RNP and template injection with and without co-injection of the xrcc6-targeting PMO. For ripor2, possibly tp53, but not ush2a, an increase in knock-in events is observed after co-injection of sgRNA-Cas9 RNPs, template oligonucleotide and the xrcc6-targeting PMO. Results are expressed as mean (bars) and individual replicates (scatterplot). Only reads that include the PAM variant are included in the quantification to ensure that all quantified correct knock-in reads are the project of homology directed repair. (C) Comparison of changes in knock-in efficiency between embryos of the same clutch of eggs. For ush2a C771F RNPs, addition of the PMO to the injection mixture leads to variable outcomes in knock-in efficiency between the replicate injections. Three out of the four replicate injections of ripor2 del12 RNPs with xrcc6-tarageting PMO display a higher knock-in efficiency, while for tp53 the addition of the PMO leads to an increase in knock-in efficiency in one of the two replicate injections. |