|
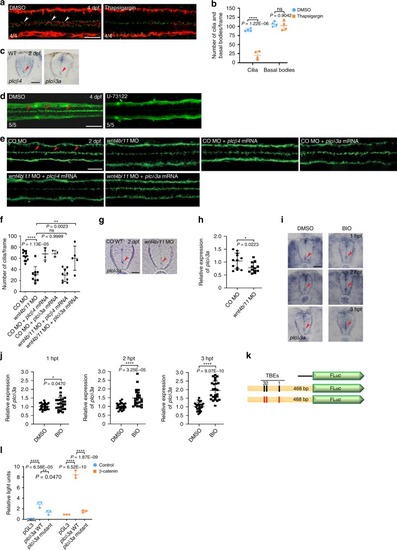
<italic>plcδ3a</italic> is a target gene of Wnt signaling and plays an important role in the maintenance of ependymal motile cilia.a DMSO (vehicle control) or thapsigargin (4 nl at 5 μM) was microinjected into the hindbrain ventricles of zebrafish larvae at 4 dpf and the larvae were double immunostained with anti-acetylated α-tubulin antibody (red) and anti-γ-tubulin antibody (green) 4 h after injection. Arrowheads indicate motile cilia. Dorsal view anterior to the left. Scale bar = 20 μm. b Quantification of the number of cilia and basal bodies per frame in embryos in a. Mean ± SD. ****P < 0.0001 by two-tailed unpaired Student’s t test (n = 4 embryos per group; one frame per embryo). ns, not significant. c Embryos at 2 dpf were probed with plcβ4 or plcδ3a riboprobes and their SCs were cross-sectioned. Images are oriented ventral to the bottom. Arrowheads represent ECs. Scale bar = 20 μm. d DMSO (vehicle control) or U-73122 (4 nl at 10 μM) was microinjected into the hindbrain ventricles of zebrafish larvae at 4 dpf and the larvae were IF stained with anti-acetylated α-tubulin antibody 4 h after microinjection. Dorsal view anterior to the left. Scale bar = 20 μm. e Control morphants or wnt4b/11 double morphants were microinjected with plcβ4 or plcδ3a mRNA and immunostained with anti-acetylated α-tubulin antibody at 2 dpf. Arrowheads represent motile cilia. Dorsal view anterior to the left. Scale bar = 20 μm. f Quantification of the number of motile cilia per frame in embryos in e. Mean ± SD. **P < 0.01 and ****P < 0.0001 by one-way ANOVA with Tukey’s HSD post hoc test (control morphants: n = 12 embryos; wnt4b/11 double morphants: n = 9 embryos; control morphants + plcβ4 mRNA: n = 4 embryos; control morphants + plcδ3a mRNA: n = 3 embryos; wnt4b/11 double morphants + plcβ4 mRNA: n = 9 embryos; wnt4b/11 double morphants + plcδ3a mRNA: n = 5 embryos; one frame per embryo). ns: not significant. g Control morphants or wnt4b/11 double morphants at 2 dpf were probed with plcδ3a riboprobes and their SCs were cross-sectioned. Images are oriented ventral to the bottom. Arrowheads point to ECs. Scale bar = 20 μm. h RNAs were extracted from each group (20 embryos in g) at 2 dpf and levels of plcδ3a mRNA were assessed by qPCR. Mean ± SD. *P < 0.05 by two-tailed unpaired Student’s t test from four biological replicates (three technical replicates each). CO: Control. i Embryos at 45 hpf were treated with DMSO or BIO (5 μM) for 1–3 h and probed with plcδ3a riboprobes, and their SCs were sectioned. Images are oriented ventral to the bottom. hpt: hours post-treatment. Arrowhead indicates ECs. Scale bar = 20 μm. j Upon BIO treatment, RNAs were extracted from each group (20 embryos in (i)) and levels of plcδ3a mRNAs were assessed by qPCR. Mean ± SD. *P < 0.05 and ****P < 0.0001 by two-tailed unpaired Student’s t test from three biological replicates (eight technical replicates each). k Schematic of the plcδ3a promoter-firefly luciferase construct used in the luciferase reporter assay. Black rectangles represent three (1–3) Tcf binding elements (TBEs) and red rectangles TBEs with deletions. l HEK 293T cells were transfected with WT or mutant plcδ3a promoter-firefly luciferase constructs and Renilla luciferase plasmid with or without β-catenin plasmid, and processed for dual luciferase assay. Relative Light Units: firefly luciferase activity/Renilla luciferase activity. **P < 0.01 and ****P < 0.0001 by one-way ANOVA with Tukey’s HSD post hoc test (n = 3 culture replicates per group; each culture was assayed three times).
|