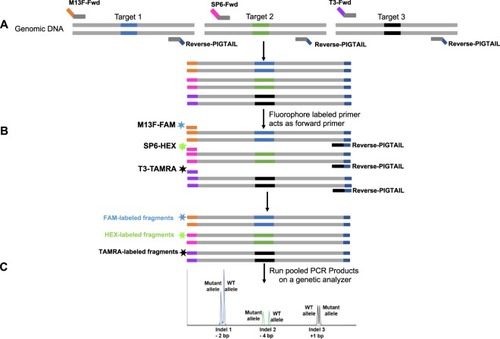
Overview of MultiFRAGing method. (A) Primer design strategy for multiple targets. Gene-specific primers are designed to generate 180–300 bp fragments. The gene-specific forward primers are attached with an adapter sequence (M13Fwd, SP6, and T3), and the reverse primer contains a short PIGTAIL sequence to suppress the stutter peaks. Tailed gene-specific primers add M13Fwd or SP6 or T3 adapter sequences in the first few PCR cycles. (B) A third primer with same adapter sequence attached to a fluorescent dye (FAM, HEX and TAMRA) is used to generated fluorescently labeled fragments. After few PCR cycles, fluorophore-labeled primers act as forward primer and bind to the respective adapter sequences. In the subsequent PCR cycles, most fragments will incorporate fluorophore thus generating double-stranded fluorescent fragments. Multiple fragments are generated in a single PCR reaction. These fragments can either be tagged with same dye and generate products of different size or tagged with different dyes. (C) Pooled PCR products are mixed with a size standard to run on a genetic analyzer. Fragments sizes are plotted, and indel size can be measured based on the expected size of the wild type fragment. Wild-type samples will have one size (allele), while heterozygous samples will show two sizes (alleles).
|

