Fig. 5
- ID
- ZDB-FIG-190723-92
- Publication
- Sasaki et al., 2019 - Calaxin is required for cilia-driven determination of vertebrate laterality
- Other Figures
- All Figure Page
- Back to All Figure Page
|
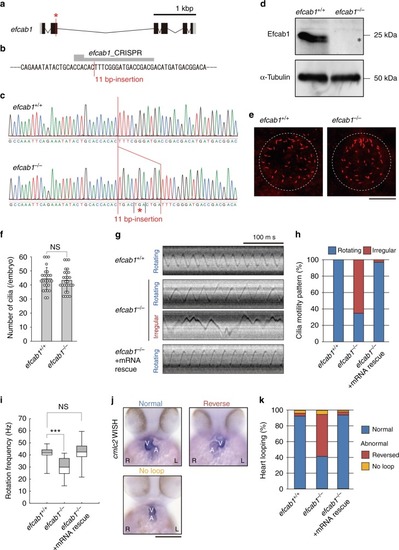
Mutation of zebrafish efcab1 causes abnormal motility of Kupffer’s vesicle cilia. a Genomic organization of zebrafish efcab1. Black boxes: exons. Gray boxes: untranslated regions. Red asterisk indicates the genome-editing target site. b CRISPR/Cas9 target sequence. c Sanger sequencing of efcab1+/+ and efcab1−/− fish around the genome-editing target site. The 11 bp-insertion in efcab1−/− includes a stop codon (red asterisk). d Immunoblot of testis lysate. The induced mutation deleted Efcab1 (asterisk). α-tubulin: loading control. e Kupffer’s vesicle cilia were visualized by immunofluorescence staining with acetylated-tubulin antibodies. f Measurement of the number of Kupffer’s vesicle cilia showed no significant differences between efcab1+/+ and efcab1−/− fish. N (embryo) = 26 (efcab1+/+) and 27 (efcab1−/−). Values indicate mean ± SD. g Typical kymographs of Kupffer’s vesicle cilia in efcab1+/+, efcab1−/−, and mRNA-rescued efcab1−/− fish. Kymograph patterns were categorized into two classes: rotating (blue) and irregular (red). h Ratios of each motility class. N (cilia) = 55 (efcab1+/+), 72 (efcab1−/−) and 65 (mRNA-rescued efcab1−/−). i Rotational frequencies of Kupffer’s vesicle cilia. Boxes correspond to the first and third quartiles, and whiskers extend to the full range of the data. N (cilia) = 55 (efcab1+/+), 25 (efcab1−/−) and 63 (mRNA-rescued efcab1−/−). ***p < 0.001 vs. efcab1+/+ (Student’s t-test). j Ventral views of 48 hpf embryos. Heart looping was visualized by whole-mount in situ hybridization of cmlc2. L, left; R, right; V, ventricle; A, atrium. k Directions of heart looping. N (embryo) = 108 (efcab1+/+), 114 (efcab1−/−), and 51 (mRNA-rescued efcab1−/−) |
| Fish: | |
|---|---|
| Observed In: | |
| Stage Range: | 5-9 somites to Long-pec |