Fig. 6
- ID
- ZDB-FIG-150915-2
- Publication
- Varshney et al., 2015 - High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9
- Other Figures
- All Figure Page
- Back to All Figure Page
|
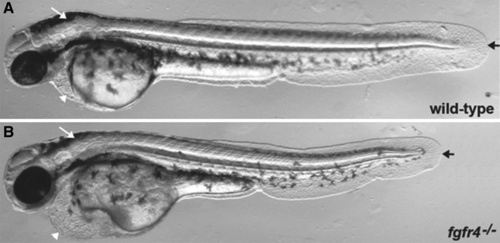
fgfr4-/- phenotype identified by inbreeding injected founder fish. Two single-guide RNAs (sgRNAs) targeting two different exons of the fgfr4 gene were co-injected with Cas9mRNA into wild-type embryos and raised to adults to generate founder fish (F0). Six pairs of founder fish (F0) were inbred and scored for mutant phenotypes. One of the six pairs showed multiple embryos with the same phenotype observable at 48 h post-fertilization. (A) Wild-type embryo at 48 h post-fertilization. White arrow indicates a normally sized hindbrain, white arrowhead indicates a normal heart chamber, and black arrow represents a normal tail length. (B) An fgfr4 compound heterozygous mutation displaying multiple phenotypes. White arrow indicates a reduced hindbrain region, white arrowhead points to a heart edema, and black arrow shows the shortened body axis. All phenotypes were only found in the compound heterozygous mutant embryos. |
| Fish: | |
|---|---|
| Knockdown Reagents: | |
| Observed In: | |
| Stage: | Long-pec |