Fig. 6
|
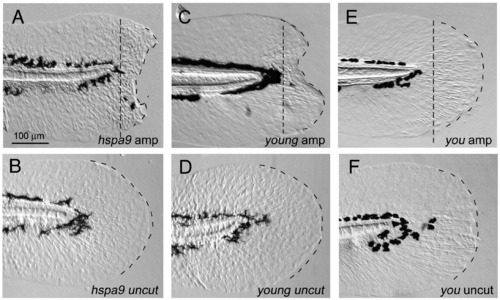
Genetic mutations affecting the larval fin fold regeneration. (A, B) Disrupted larval fin fold regeneration in hspa9 mutants. The larval regeneration was apparently impaired (severe defect in 44/56 mutants, mild defect in 8/56 mutants; (A), whereas the uncut mutants did not show such defects (B). (C, D) Disrupted larval fin fold regeneration in yng/smarca4 mutants. A complete penetrance of defect in regeneration was observed in yng/smarca4 mutants (n = 28/28 mutants, C). Despite the mutant phenotypes such as smaller eyes and curved body axis, hspa9 and yng/smarca4 mutants were alive at 5 dpf and retain a good cellular viability. (E, F) Normal regeneration in other mutants. Several other mutants including you mutants, which have a defect in hedgehog signaling (Kawakami et al., 2005), did not display such defects (n = 15, E). The same magnifications for panels A?F (scale bar in panel A). |
| Fish: | |
|---|---|
| Observed In: | |
| Stage: | Prim-5 |
Reprinted from Developmental Biology, 325(1), Yoshinari, N., Ishida, T., Kudo, A., and Kawakami, A., Gene expression and functional analysis of zebrafish larval fin fold regeneration, 71-81, Copyright (2009) with permission from Elsevier. Full text @ Dev. Biol.