- Title
-
Incoherent collective cell chemotaxis underlies organ dysmorphia in a model of branchio-oto-renal syndrome
- Authors
- Borges, A., Pinto-Teixeira, F., Wibowo, I., Pogoda, H.M., Hammerschmidt, M., Kawakami, K., López-Schier, H., Miranda-Rodríguez, J.R.
- Source
- Full text @ MicroPubl Biol
|
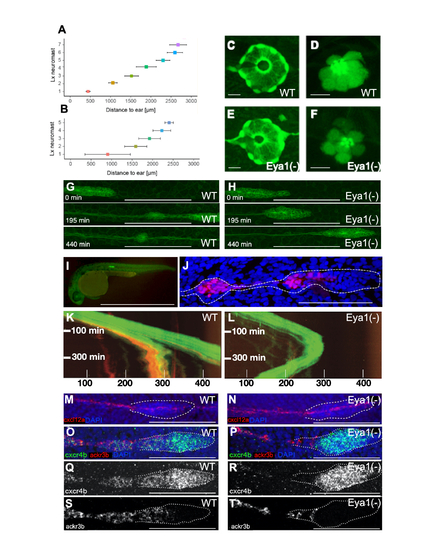
Loss of eya1 disrupts lateral-line development: (A,B) Plot of the distance (in ?m) between the caudal limit of the otic vesicle and the average number of deposited neuromasts in wild-type (E) and eya1-mutant (F) specimens at 3 dpf (mean ± s.d.). N= 4 for wild type and N=9 for eya1-/-. (C-F) Live images of neuromasts of the transgenic lines Et(krt4:EGFP)sqet20 (C,E) and Et(krt4:EGFP)sqet4 (D,F) at 6dpf in wild-type (C,D), eya1 mutants (E,F) revealing supporting cells (C,E) and hair cells (D,F). (G,H) High magnification confocal images of a wild type (G) and mutant (H) primordium. At 195 minutes of migration, the wild-type primordium has deposited one pro-neuromast, whereas the eya1(-) primordium failed to do so even after 440 minutes. (I) A transgenic gSAG181A larva at 30hpf. This line specifically expresses EGFP in the posterior lateral line primordium. (J) The primordium and a pro-neuromast in a SAGFF(LF)19A;UAS:RFP larva. SAGFF(LF)19A expresses a Gal4 protein in rear primordial cells, pro-neuromasts and inter-neuromast cells. (K) Kymograph from a time-lapse movie focusing on the migrating primordium. The trailing RFP signal driven by SAGFF(LF)19A signal is associated with pro-neuromast rosettogenesis. A few cells expressing 19A:RFP remain in the trailing edge after pro-neuromast deposition. The leading edge advances linearly with a velocity of roughly 80 microns per hour at 28°C. (L) Kymograph from a representative eya1 Crispant larva in the 181A;19A:RFP transgenic background. No neuromast deposition is seen for the duration of the movie. The fish was confirmed to express 19A:RFP in older neuromasts but barely any red fluorescence is seen in the trailing edge of the primordium. The curved shape of the trajectory indicates that after stalling, the primordium performs a U-turn and starts backward migration. Black gaps in the solid green band reflect transient splitting events of the primordium. Units on the x-axis are micrometers (?m). (M-T) Representative fluorescent whole-mount in situ hybridizations (M,N) of cxcl12a (red), counterstained with DAPI (blue) to reveal the nuclei for better identification of the primordium (white dotted outline). It shows that the cxcl12a gene is expressed along the horizontal myoseptum in the wild type (M) and eya1 mutants (N). (O-T) cxcr4b (green) and ackr3b (red) gene-expression profiles in wild type (O,Q,S) and eya1 mutants (P,R,T) The cxcr4b gene is strongly expressed in the leading region of primordium in both in wild-type and eya1 mutants (O-R). The expression of ackr3b, however, is strong in the trailing region of the wild-type primordium (overlapping with cxcr4b) (O,Q,S), but almost completely lost in eya1 mutants, as it is restricted to the very end of the trailing region and never overlaps with cxcr4b (P,R,T). HCR conducted in al least 10 samples. Scale bars: C-F 10 ?m; G-H 100 ?m; I 1mm; J, 100 ?m; M-T 100 ?m. |