- Title
-
Leukotriene B4 receptor 2 governs macrophage migration during tissue inflammation
- Authors
- Ermis, E., Nargis, T., Webster, K., Tersey, S.A., Anderson, R.M., Mirmira, R.G.
- Source
- Full text @ J. Biol. Chem.
|
|
|
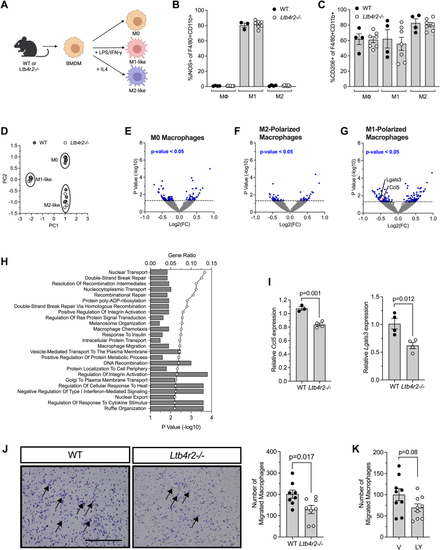
EXPRESSION / LABELING:
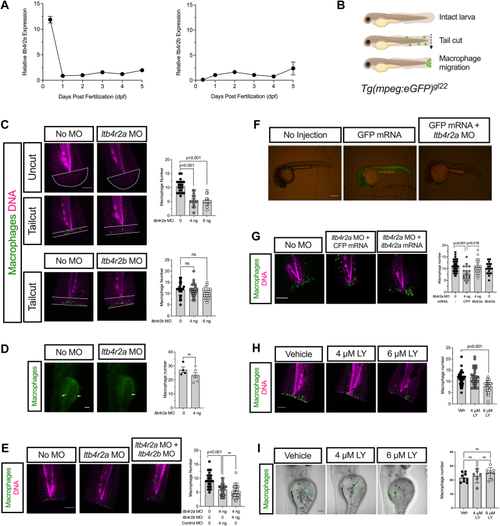
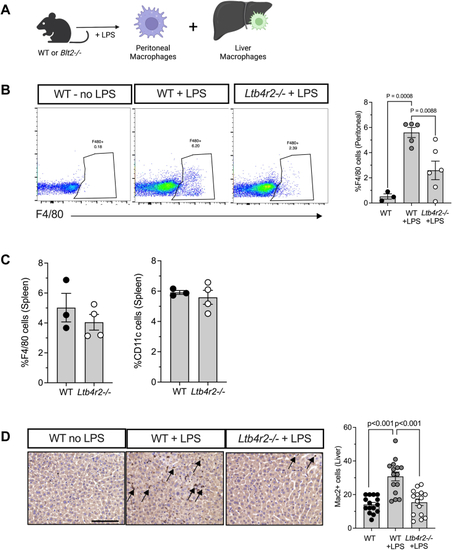
PHENOTYPE:
|
|
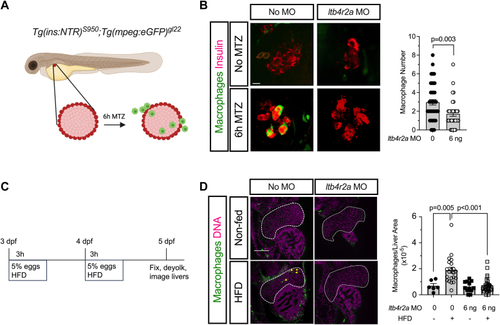
PHENOTYPE:
|
|
|