- Title
-
Cdx1b protects intestinal cell fate by repressing signaling networks for liver specification
- Authors
- Jin, Q., Gao, Y., Shuai, S., Chen, Y., Wang, K., Chen, J., Peng, J., Gao, C.
- Source
- Full text @ J. Genet. Genomics
|
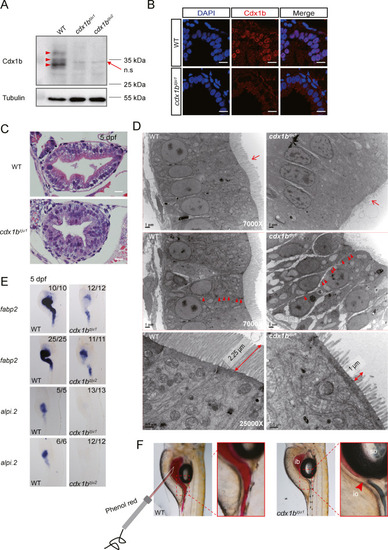
Fig. 1. Cdx1b is essential for the development of a functional intestine in zebrafish. A: Western blot showing the depletion of Cdx1b protein in two cdx1b mutants cdx1bzju1/zju1 (cdx1bzju1) and cdx1bzju2/zju2 (cdx1bzju2) compared with WT at 5 dpf. In WT, Cdx1b showed three isoforms (red arrow head). B: Immunostaining of Cdx1b (red) in the intestines of WT and cdx1bzju1/zju1. Cdx1b is a nuclear localized protein. DAPI stains the nuclei. Scale bar, 10 ?m. C: HE staining showing the disorganized gut epithelium in cdx1bzju1/zju1 compared with WT at 5 dpf. Scale bar, 10 ?m. D: TEM images showing the shortened microvilli and loosened cell-cell junction in cdx1bzju1/zju1 compared with WT at 5 dpf. Red arrow, microvilli; double headed arrow, length of microvilli; arrow head, cell-cell junction. Scale bar, 1 ?m or 0.5 ?m (bottom two images). E: WISH showing the decreased expression of fabp2 and near absence of alpi.2 in the cdx1bzju1/zju1 and cdx1bzju2/zju2 intestine compared with that in WT at 5 dpf. F: Images showing the blockage of intestinal fluid by visualizing the injected phenol-red at the junction between the intestinal bulb (ib) and mid-intestine in cdx1bzju1/zju1 compared with WT. n.s, non-specific band. io, intestinal obstruction; sb, swim bladder. |
|
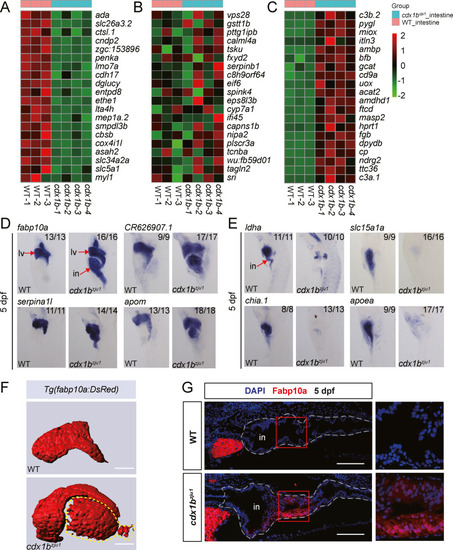
Fig. 2. Cdx1b depletion leads to ectopic expression of a cohort of liver-specific genes in the intestine. A?C: Heat maps showing the representative 20 downregulated intestine-specific genes (A), 20 unchanged intestine-specific genes (B), and 20 upregulated liver-specific genes (C) in four cdx1bzju1/zju1 samples (cdx1b) identified by RNA-seq. RNA-seq data for the three WT samples were adopted from our recently published report (Gao et al., 2022). D and E: WISH showing the upregulation of four liver-specific genes (fabp10a, serpina1l, CR626907.1, and apom) (D) and down-regulation of four intestine-specific genes (ldha, chia.1, slc15a1a, and apoea) (E) in the cdx1bzju1/zju1 intestine compared with WT at 5 dpf. F: Representative 3-D images of the DsRed fluorescence in WT and cdx1bzju1/zju1 in the Tg(fabp10a:DsRed) background at 5 dpf. The ectopic expression of RFP in the cdx1bzju1/zju1 intestine is outlined by a dashed yellow line. G: Immunostaining showing the ectopic expression of Fabp10a in the cdx1bzju1/zju1 intestine compared with WT. The Fabp10a-positive signals appeared to be limited in the posterior region of the bulb and anterior region of the mid-intestine. The intestine is outlined with a white dashed line. Red frame, enlarged view on the right. Scale bar, 100 ?m. lv, liver; in, intestine. |
|
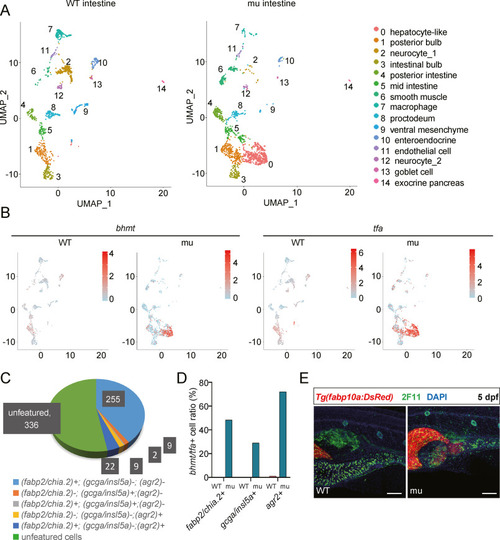
Fig. 3. Depletion of Cdx1b leads to ectopic hepatic gene expression in three major types of gut epithelia cells. A: Visualization of integrated unsupervised clustering of cells in UMAP plot of the sequenced 1229 cells from the WT intestines (Gao et al., 2022) (left UMAP panel) and 1548 cells from cdx1bzju1/zju1 (mu) intestines (right UMAP panel) at 5 dpf. Annotation of the 15 cell types is shown on the right. B: Feature plot showing the high expression of bhmt and tfa in the group of hepatocyte-like cells in cdx1bzju1/zju1 (mu) which was not present in WT. C: Colored pie-chart showing the proportion of bhmt+/tfa+ hepatocyte-like cells (total 633 cells) co-expressing marker genes of enterocytes (fabp2+/chia.2+), enteroendocrine cells (gcga+/insl5a+) and goblet cells (agr2+) and others. Corresponding number of cells are provided in the dark grey box. D: Histogram showing the percentage of bhmt+/tfa+ in the group of fabp2+/chia.2+, gcga+/insl5a+ and agr2+ cells in WT and cdx1bzju1 (mu), respectively. Note that no bhmt+/tfa+ cell was detected in WT. E: Whole-mount immunostaining showing that only a proportion of the ectopically expressed RFP (red) was co-expressed with 2F11 (green) in the cdx1bzju1 intestine which was not observed in WT in the Tg(fabp10a:DsRed) background at 5 dpf. 2F11 staining marks the goblet cells, enteroendocrine cells, biliary cells, and swimming bladder. DAPI stains the nuclei. Scale bar, 100 ?m. |
|
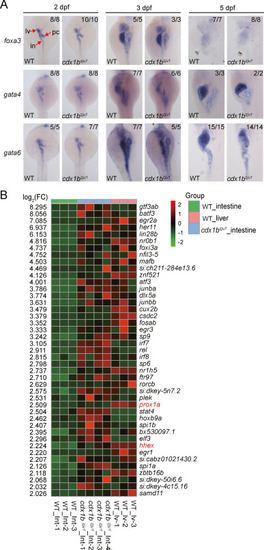
Fig. 4. Depletion of Cdx1b does not obviously affect the endoderm patterning. A: Patterning of the endoderm-derived organs was not visually altered in cdx1bzju1/zju1 at 2 dpf, 3 dpf, and 5 dpf as revealed by WISH using the pan-endodermal markers foxa3, gata4, and gata6 probes. B: Heatmap showing 43 TFs whose expressions were upregulated in 4 cdx1bzju1/zju1 intestine samples (cdx1bzju1/zju1-Int) compared with that in three WT intestine samples (wt_Int) (cut-off value: log2[fold-change] > 2, P < 0.01). Heatmap also shows the expression of these TFs in three WT liver (WT_lv) at 5 dpf. RNA-seq data for three WT_Int and three WT_lv samples were adopted from our recently published report (Gao et al., 2022). |
|
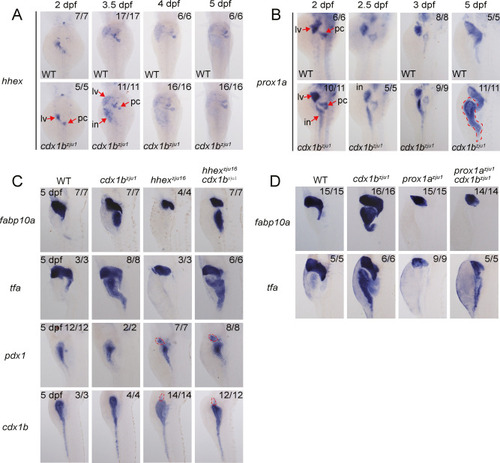
Fig. 5. Ectopically expressed hhex and prox1a in the cdx1bzju1/zju1 intestine are partially responsible for the ectopic expression of fabp0a and tfa. A and B: WISH showing the intestinal ectopic expression of prox1a (A) and hhex (B) in cdx1bzju1/zju1 but not in WT at different developmental stages as indicated. C: WISH comparing the expression patterns of tfa, fabp10a, pdx1, and cdx1b in WT, cdx1bzju1/zju1 single, hhexzju16/zju16 single and hhexzju16/zju16 cdx1bzju1/zju1 double mutants at 5 dpf. Red circle: ectopic expression of cdx1b and pdx1 in the hhexzju16/zju16 background. D: WISH showing the expression of fabp10a and tfa in WT, prox1azju1/zju1 single, cdx1bzju1/zju1 single, and prox1azju1/zju1 cdx1bzju1/zju1 double mutant at 5 dpf. Numbers on the right corner: number of embryos exhibiting the expression pattern (numerator) over number of embryos examined. lv, liver; in, intestine; pc, pancreas. |
|
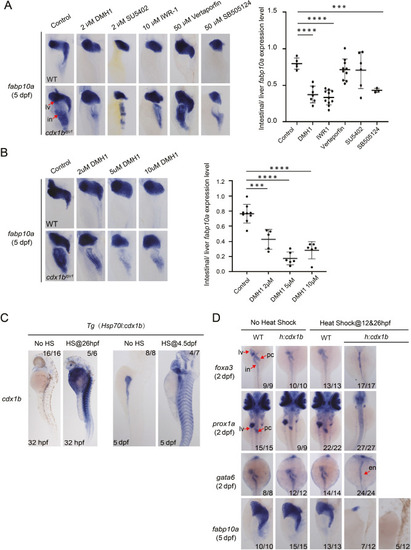
Fig. 6. Cdx1b represses multiple signaling pathways essential for liver development. A: The effect of various signaling inhibitors on the hepatized intestine phenotype in cdx1bzju1/zju1. Left, representative images showing the effect of the Bmp-signaling inhibitor DMH1, Fgf-signaling inhibitor SU5402, Wnt-signaling inhibitor IWR-1, Hippo-signaling inhibitor Verteporfin and TGF?-signaling inhibitor SB505124 on the intestinal ectopic expression of fabp10a in WT (top row) and cdx1bzju1/zju1 (lower row). Right, the intestine versus liver ratio of the fabp10a signals in cdx1bzju1/zju1. B: The mitigation effect of Bmp inhibitor DMH1 on the hepatized intestine phenotype is dosage dependent. Left, representative WISH images; right, the intestine versus liver ratio of the fabp10a signals in cdx1bzju1/zju1. In (A) and (B), embryos were treated with inhibitors starting from 36 hpf, respectively, and were subjected to WISH at 5 dpf with the fabp10a probe. Each spot represented one embryo. ???, P < 0.001; ????, P < 0.0001. C and D: Cdx1b overexpression represses liver organogenesis. In (C), WISH showing the cdx1b expression in the Tg(hsp70l:cdx1b) embryos treated with or without heat shock. Left two panels, WISH at 32 hpf after heat shock treatment (HS) at 26 hpf for 30 min; Right two panels, WISH at 5 dpf after heat shock treatment at 4.5 dpf for 30 min. In (D), WISH comparing the expression patterns of foxa3, prox1a and gata6 at 2 dpf, and fabp10a at 5 dpf between WT and Tg(Hsp70:cdx1b) (h:cdx1b) embryos treated with or without heat shock. For heat shock samples, embryos were heat-shocked at 12 hpf and 26 hpf two time points, each for 30 min, and were then collected for WISH at 2 dpf or 5 dpf. Numbers on the right corner, number of embryos exhibiting the expression pattern (numerator) over number of embryos examined. en, endoderm; in, intestine; lv, liver; in, intestine; pc, pancreas. |
Reprinted from Journal of genetics and genomics = Yi chuan xue bao, 49(12), Jin, Q., Gao, Y., Shuai, S., Chen, Y., Wang, K., Chen, J., Peng, J., Gao, C., Cdx1b protects intestinal cell fate by repressing signaling networks for liver specification, 1101-1113, Copyright (2022) with permission from Elsevier. Full text @ J. Genet. Genomics