- Title
-
Swift Large-scale Examination of Directed Genome Editing
- Authors
- Hammouda, O.T., Böttger, F., Wittbrodt, J., Thumberger, T.
- Source
- Full text @ PLoS One
|
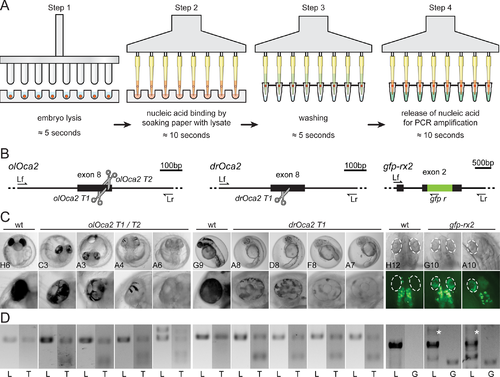
Application of filter-in-tips in high throughput, SLEDGE hammer protocol. A) Schematic workflow of high-throughput 96-well plate based SLEDGE hammer protocol with filter-in-tips. Step 1 lysis: (96-pin) mortar used to simultaneously grind individual embryos in fin-clip buffer. Step 2 binding: using a multichannel pipet equipped with filter-in-tips, pipet up lysate to let nucleic acids (red) bind to cellulose filter discs (soak for ≈10 sec). Release lysate back into wells for storage. Step 3 washing: wash filter discs containing nucleic acids (red) by pipetting nuclease free water in and out (≈5 sec). Step 4 elution: pipet up pre-mixed PCR mixture (wait ≈10 sec) to release nucleic acids and pipet back for amplification. Note: entire procedure takes less than 10 minutes (6 minutes processing time). B) Schematic representation of CRISPR/Cas9 mediated NHEJ-based knock-out (medaka oca2, olOca2; zebrafish oca2, drOca2) and HDR-mediated single-copy integration of gfp sequence in frame with medaka rx2 (cf. Gutierrez et al., 2018 [20]). Location of sgRNA target sites shown by scissors. Primers used for analysis indicated; locus forward (Lf), locus reverse (Lr) of the respective gene. rx2 locus shown after single-copy HDR-mediated integration of gfp. C) Besides fully pigmented eyes in wild-type (wt) medaka and zebrafish, varying degrees of pigment loss in body and RPE (blow-up) of representative crispants injected with individual sgRNAs targeting oca2 exon 8. Retinae (dashed ellipses) of injected embryos show GFP expressing cells after HDR/donor mediated integration of gfp into rx2 open reading frame. Note: unspecific autofluorescence of body pigment. 96-well plate coordinates of specimens indicated (see S2 Fig). D) Genotyping of representative embryos in C. Locus PCR (L; primers Lf/Lr) of respective genes and T7EI assay (T) validating indel formation in oca2 loci. In gfp-rx2 tagging, besides non-gfp-integrated locus band (L; primers Lf/Lr), single gfp integration evident by PCR (white asterisk). Integration validated by locus-gfp band (G; primers Lf/gfp r).
|