- Title
-
Ribozyme Mediated gRNA Generation for In Vitro and In Vivo CRISPR/Cas9 Mutagenesis
- Authors
- Lee, R.T., Ng, A.S., Ingham, P.W.
- Source
- Full text @ PLoS One
|
gRNA generated containing HH and HDV ribozyme is efficient at inducing mutagenesis of its target site. (A) mCherryHH-gRNA-HDV construct used to generate gRNA. H2a mCherry is used to label cells expressing gRNA and MALAT is used to stabilize the RNA after cleavage by HH and HDV ribozyme. (B) Injection of Smo gRNA together with Cas9 mRNA causes mutation of its target sequence. (C) Images of smyhc1: GFP embryos at 2dpf show that the majority of GFP expressing cells are lost upon injection of GFP gRNA and Cas9 mRNA. On average, there are ~24 GFP positive slow muscle fibers per somite in uninjected smyhc1: GFP embryos whereas only 6 GFP positive slow muscle fibres remained in each somite of injected smyhc1: GFP embryos (n = 5). Scale bars: 40 ?m. |
|
RNAPol II dependent promoters can be used to generate gRNA in-vivo and induce mutagenesis. (A) DNA Construct containing Cas9 and gRNA targeting GFP, both driven by ubiquitin promoter. (B) Transient expression of this construct in smyhc1:GFP embryos at 5dpf shows that some cells normally expressing GFP are lost. Cells expressing construct are labelled with H2a mCherry. On average 14 GFP positive slow muscle fibres remained in each somite after injections with this construct (n = 6), whereas there are ~24 GFP positive slow muscle fibers per somite in smyhc1: GFP embryos. Scale bars: 40 ?m. (C) PAGE analysis of individual embryos injected with Ubi: Cas9 Ubi: GFPgRNA (left) and RNA of Cas9/GFP gRNA (right) show that GFP is mutagenized. (D) A heat shock promoter was used to drive the expression of two gRNA against Smo 5? UTR. This construct was used to generate transgenic zebrafish (HSP: Smo gRNA). (E) Curved embryos seen after injection of AB with Cas9 mRNA and Smo gRNA targeting smo 5? UTR and smo ATG (42 out of 117); and HSP: Smo gRNA embryos injected with Cas9 mRNA after heat shock (46 out of 131). Scale bars: 500 ?m. (F) PAGE analysis of embryos from control (uninjected HSP: Smo gRNA) shows that Smo 5? UTR and Smo ATG is mutagenized. Furthermore deletion of the region between the two gRNA targets can be detected (red asterisk). |
|
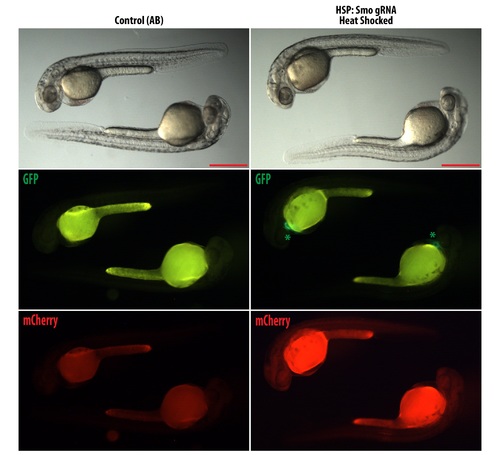
HSP: Smo gRNA express mCherry after heat shock. Embryos were heat shocked according to materials and methods section to test for expression of mCherry. Left panel show wild type embryos and right panel show HSP: Smo gRNA. Green asterisk show the expression of GFP in the heart from the cmlc: GFP used as the transgenesis marker. Embryos shown are 30 hpf. Scale bars: 500 ?m. |
|
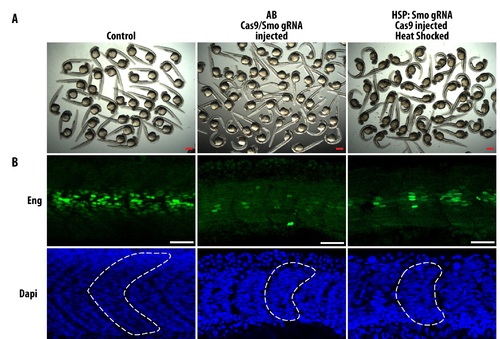
Characterization of Smo gRNA injected and HSP: Smo gRNA mutants. (A) Overview of control embryos (uninjected HSP: Smo gRNA), embryos injected with Cas9 mRNA and Smo gRNA, and heat shocked HSP: Smo gRNA embryos injected with Cas9 mRNA. Scale bars: 500 ?m. (B) Embryos were stained with Engrailed (Eng) to show defects in muscle specification when smo is mutagenesized. In the middle and right most panel, curved embryos were imaged and these embryos have defective formation of muscle pioneers and media fast fibers which are dependent on smo for its formation (Embryos shown are representative of the variation in observed phenotypes). Furthermore, these embryos have U-shaped somites (dotted line showing outline of somite in DAPI images) typical of lost of hedgehog signalling. Scale bars: 40 ?m. |