- Title
-
Hypocretin neuron-specific transcriptome profiling identifies the sleep modulator Kcnh4a
- Authors
- Yelin-Bekerman, L., Elbaz, I., Diber, A., Dahary, D., Gibbs-Bar, L., Alon, S., Lerer-Goldshtein, T., Appelbaum, L.
- Source
- Full text @ Elife
|
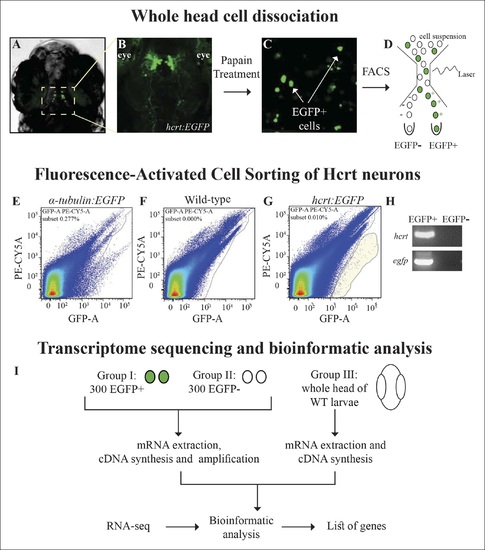
Isolation of Hcrt neurons, RNA-seq, and experimental design. (A) Dorsal view of 6 dpf hcrt:EGFP larvae. (B) Dorsal view of the hypothalamus region of 6 dpf hcrt:EGFP larvae expressing EGFP in Hcrt neurons. (C) Cell suspension from the whole head of 6 dpf hcrt:EGFP larvae. (D?G) The cells were sorted based on size and fluorescence intensity. The fluorescence thresholds (gray curve) were set based on larvae expressing EGFP under the control of α-tubulin promoter (positive control) (E) and WT larvae (negative control) (F). Positive EGFP cells (EGFP+) sorted from hcrt:EGFP larvae are marked with gray shade (G). (H) PCR amplification of hcrt and egfp was performed on cDNA synthesized from EGFP+ and EGFP- cells sorted from hcrt:EGFP larvae. (I) FAC-sorting yielded two groups of cells: Group I containing EGFP+ and Group II containing EGFP- cells. A third group contained cells from whole head of WT larvae. The cDNA of groups I and II was amplified and the three groups were then subjected to RNA-seq and bioinformatic analysis to obtain a list of Hcrt-neuron?enriched genes. |
|
The expression pattern of selected candidate Hcrt-neuron?specific genes. (A) Table presenting the top 20 Hcrt-enriched transcripts. (B?M) Dorsal view of whole-mount ISH-stained 2 dpf WT larvae. Based on the RNA-seq and the bioinformatic analysis, the expression pattern of selected candidate Hcrt-neuron?specific genes was determined. The expression pattern of hcrt (B) was used for comparison. EXPRESSION / LABELING:
|
|
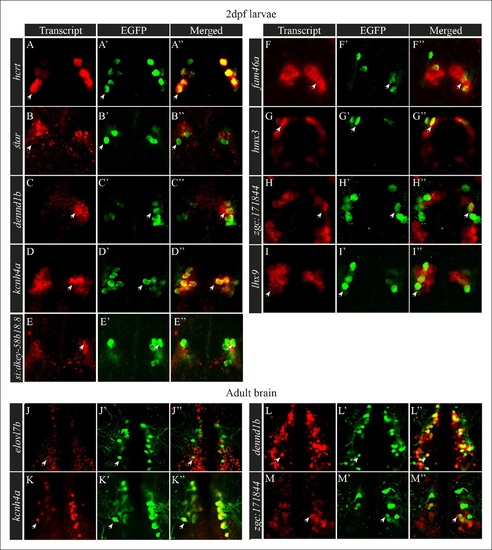
Selected candidate genes are expressed in Hcrt neurons. (A?I′′) Double fluorescent staining of the candidate genes (red) and EGFP (green) was performed in 2 dpf hcrt:EGFP larvae using whole-mount ISH and immunofluorescence, respectively. White arrows indicate representative co-expressing cells. All confocal images show single plane view of 0.5 �M width. (J?M′′) Double fluorescent ISH and immunofluorescence experiments in brain sections of hcrt:EGFP adult zebrafish. Co-localization of candidate genes (red) and EGFP (green) in Hcrt neurons is shown. All images show single plane view of 0.5 �M width. |
|
Candidate genes are expressed in cell populations located adjacent to Hcrt neurons. (A?I′′) Fluorescent ISH and immunofluorescence experiments in 2 dpf larvae and adult hcrt:EGFP zebrafish showing three candidate genes that are expressed adjacently to Hcrt neurons within the hypothalamus. (A?A′′, D?D′′, G?G′′) Dorsal view of the heads showing the whole expression pattern of the genes in 40 �M z-stack. (B?B′′, E?E′′, H?H) Dorsal view of single 0.5 �M plane in 2 dpf larvae. (C?C′′, F?F′′, I?I′′) Dorsal view of single 0.5 �M plane in adult brain section. |
|
Predicted TFs that regulate the expression of Hcrt-neuron?specific transcripts. (A) TFs with a p <0.005 and their target Hcrt-neuron?specific genes. For each of these transcription factors, a combined score for each gene is calculated according to all of the predicted binding sites in its promoter. Therefore, a gene with an overrepresented binding site of a TF will have a high score for that TF and a lower p value. (B?C′′) Double fluorescent ISH of hsf1 and immunofluorescence staining of EGFP in hcrt:EGFP adult brain section. Single plane (0.5 �M width) view of the Hcrt-neuron region (C?C′′). Arrows mark representative EGFP and hsf1 co-expressing cell. EXPRESSION / LABELING:
|
|
The genomic location, phylogenetic reconstruction and structure of kcnh4a, and the generation of kcnh4a-/- zebrafish. (A) Synteny analysis shows similar genomic context of hcrt in zebrafish and mammals. Notably, kcnh4a is located a few kbs downstream to hcrt in zebrafish and mammals. (B) The 16-exon kcnh4a gene (black box = exon, white box = UTR) encodes for a voltage-gated potassium channel that includes an N-terminal chain (black bar), pore and voltage-sensing domains (S1-6, grey bar), and the C-terminal chain (red bar). (C) A cladogram-style phylogenetic tree depicting the evolutionary conservation of Kcnh4a protein among vertebrates. The tree shows topography as well as distance indicated by the branch support values above corresponding branches. (D) Generation of CRISPR-mediated kcnh4a-/- zebrafish. A 14 bp deletion was introduced in exon 5 that encodes to the S2 domain. A short mutant allele was visible on agarose gel. (E) Quantitative reverse transcription PCR shows reduction of 59% in the expression levels of kcnh4a mRNA in kcnh4a-/- 6 dpf larvae (p < 0.001). |
|
Hypothalamic kcnh4a-expressing neurons are glutamatergic. Double in-situ hybridization against kcnh4a (red, A, B) and gad67 (green, A'), or vglut2b (green, B'). All pictures are on a single optical plane of 0.5 μm. EXPRESSION / LABELING:
|
|
Sleep time and quality are reduced in kcnh4a-/- larvae during the night. (A) The locomotor activity of kcnh4a-/- (n=85), kcnh4a+/- (n=208), and kcnh4a+/+ (n=98) is shown. kcnh4a-/- larvae exhibit increased locomotor activity compared with kcnh4a+/- and kcnh4a / under LD conditions. (B) kcnh4a-/- larvae showed a significant reduction in sleep time compared with kcnh4a+/- and kcnh4a+/+ during the night. Bar charts represent the average total locomotor activity (A′) and sleep time (B′) for each genotype. Values are represented as means � SEM. (C,D) The number of sleep\wake transitions (C) and the length of sleep bout (D) are decreased in kcnh4a-/- larvae during the night. Recording of locomotor activity and sleep was performed in 6 dpf larvae continuously during 24 hr under a 14 hr light/10 hr dark cycle (white and black bars represent light and dark periods, respectively, *p<0.05, **p<0.01, ***p<0.0001, with repeated measures of ANOVA). PHENOTYPE:
|

Unillustrated author statements |