- Title
-
Expression of the transcription factor Olig2 in proliferating cells in the adult zebrafish telencephalon
- Authors
- März, M., Schmidt, R., Rastegar, S., and Strähle, U.
- Source
- Full text @ Dev. Dyn.
|
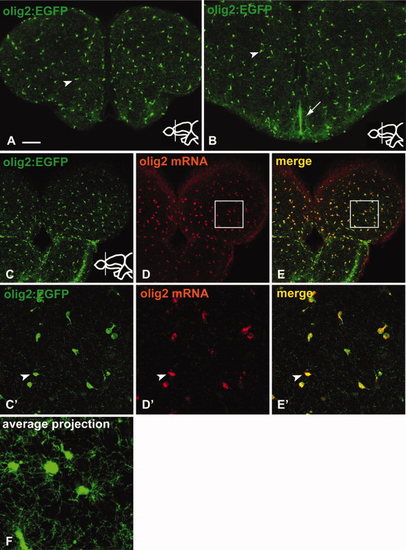
The transgenic line Tg(olig2:EGFP) represents the endogenous pattern of olig2 mRNA expression in the adult telencephalon. All views of this and all other figures are transverse sections through the telencephalon, stained with the markers indicated (colour-coded). Inserts indicate rostrocaudal positions. A, B: Immunostaining against GFP in the Tg(olig2:EGFP) transgenic line. The transgenic line expresses GFP in cells of the parenchyma (arrowhead in A, B) and at the ventricle (Vv, arrow in B). C?E: The transgenic line (C) mirrors the endogenous expression of olig2 mRNA (D) shown by fluorescent in situ hybridization and confocal microscopy. A complete overlap can be observed (E). C′?E′: High magnification of the boxed region in C?E, with an arrowhead indicating co-localization of GFP and olig2 mRNA. F: Average projection of confocal Z-stack indicating the highly branched morphology of parenchymal Tg(olig2:EGFP)-expressing cells. Scale bars = (A?E) 100 μm, (C′?E′) 25 μm, (F) 10 μm. |
|
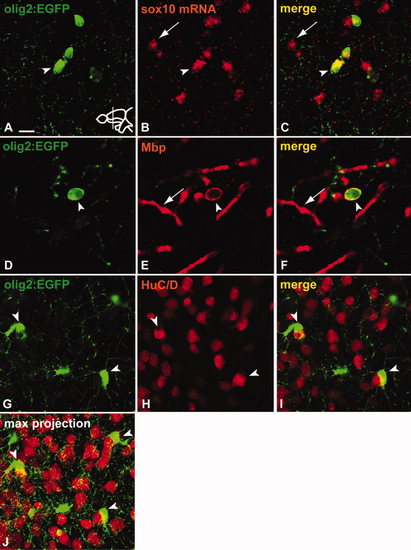
Parenchymal olig2-expressing cells belong to the oligodendrocyte lineage. A?C: Co-staining of GFP (A) with fluorescent in situ hybridization against sox10 (B). Tg(olig2:EGFP)-positive cells in the parenchyma co-express sox10 (arrowhead) (C). Arrow indicates cell expressing sox10 only. D?F: Co-staining of GFP (D) with Mbp antibody (E). Arrowhead indicates co-expression of GFP and Mbp (F), arrow indicates Mbp-positive myelin sheath. G?J:Tg(olig2:EGFP)-positive cells (G) do not express HuC/D (H) but often can be found tightly associated with HuC/D-positive neurons (arrowheads in G?J). J is a confocal Z-stack maximum projection to illustrate the tight association of Tg(olig2:EGFP)-positive cells with neighbouring neurons. Scale bars = (A?C) 10 μm, (D?F) 5 μm, (G?J) 10 μm. |
|
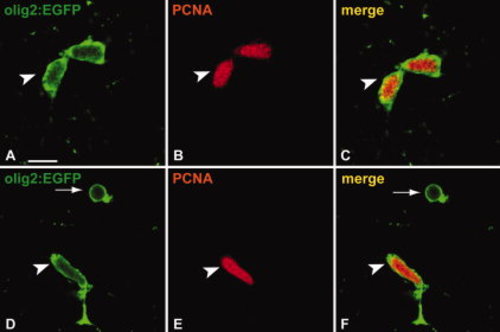
A subpopulation of Tg(olig2:EGFP)-positive cells in the parenchyma are proliferating. A?C: A subpopulation of Tg(olig2:EGFP)-positive cells (A) in the brain parenchyma express PCNA (B) and can repeatedly be found as pairs (arrowhead in C; see also arrows Fig. 4). D?F: Proliferating Tg(olig2:EGFP)-positive cells (D) in the parenchyma can display elongated nuclei reminiscent of migrating cells. Arrow indicates an Tg(olig2:EGFP)-positive/PCNA negative cell (F). Scale bar = 10 μm. |
|
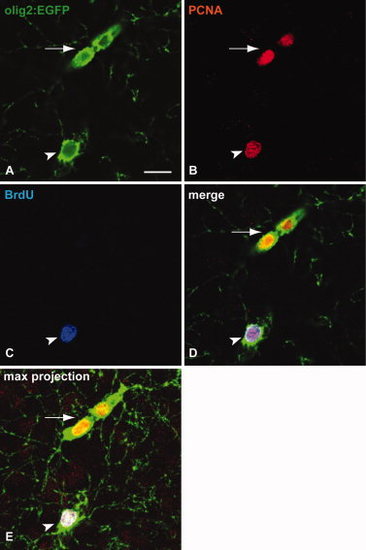
A fraction of Tg(olig2:EGFP)-positive cells are slowly proliferating cells. Animals were injected cumulatively for 5 days and kept for 4 weeks before being sacrificed and stained with the markers indicated (colour-coded). A?D: Arrowhead indicates a triple-positive cell (Tg(olig2:EGFP), PCNA, BrdU). Arrow indicates a Tg(olig2:EGFP), PCNA double-positive cell pair. E: Shows a maximum projection of a confocal Z-stack in the same region. Scale bar = 10 &mu′m. |
|
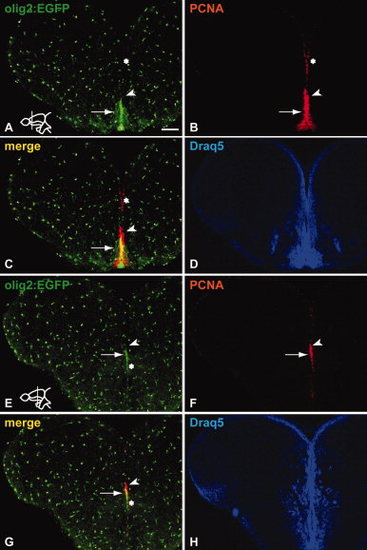
Sections through the adult zebrafish telencephalon stained immunohistochemically to reveal co-expression of Tg(olig2:EGFP) and the proliferation marker PCNA. A?D: Proliferating cells at the telencephalic ventricle are mainly Tg(olig2:EGFP) negative (asterisk in A?C). In Vv, a Tg(olig2:EGFP)-positive cell cluster in the RMS co-expresses PCNA (arrow in A?C). Arrowhead in A?C points to proliferating cells dorsal to the co-labelled region, expressing PCNA only. Draq5 staining in D and H illustrates the overall organization of nuclei in the respective sections. E?H: The ventral portion of the RMS in the medial telencephalon expresses Tg(olig2:EGFP) (arrow in E?G), while the dorsal part does not, in analogy to the RMS stripe in the anterior telencephalon (arrowhead in E?G). The asterisk indicates that the expression of Tg(olig2:EGFP) at the ventricle extends ventrally from the RMS. Scale bar = 100 μm. |
|
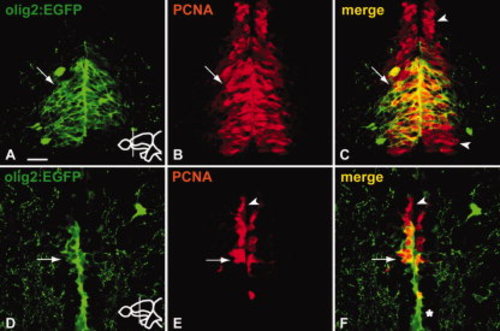
A fraction of the proliferating cells in the RMS are Tg(olig2:EGFP) positive. A?C: The majority of Tg(olig2:EGFP)-positive cells (A) in the anterior part of the RMS co-express PCNA (arrow in A?C). However, only the medial part of the anterior RMS stripe expresses Tg(olig2:EGFP). Arrowheads in C indicate PCNA-only-expressing cells located dorsal and ventral to the Tg(olig2:EGFP)-positive cluster. D?F: A part of the Tg(olig2:EGFP)-positive cells (D) in the RMS of the medial telencephalon co-express PCNA (arrow in D?F). Arrowhead in E and F indicates PCNA-only-expressing cells located dorsal to the Tg(olig2:EGFP)-positive cluster. Note that expression of Tg(olig2:EGFP) is observed also ventral to the RMS (asterisk) (see also Fig. 5E, G). Scale bar = 25 μm. |
|
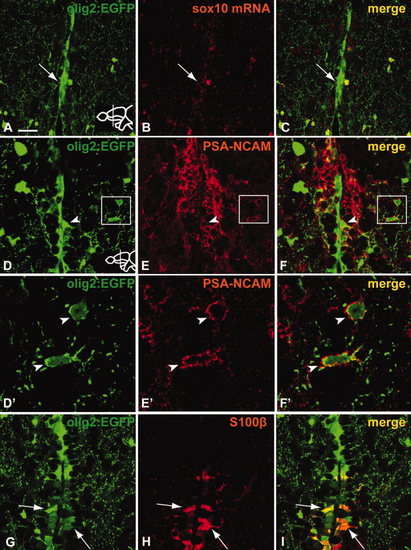
Tg(olig2:EGFP)-positive cells in the RMS do not express sox10 mRNA. A subpopulation of Tg(olig2:EGFP)-positive cells in the RMS expresses PSA-NCAM. Tg(olig2:EGFP) cells ventral to the RMS co-express the radial glia marker S100β. A?C: No or weak expression of sox10 mRNA (B) in Tg(olig2:EGFP) cells (A) of the RMS as revealed by fluorescent in situ hybridization (arrow A?C). D?F: Arrowhead in D?F indicates co-expression of Tg(olig2:EGFP) and PSA-NCAM. Boxed regions are shown in high magnification in D′?F′. D′?F′: High magnification of boxed regions in D?F. In a few cases, cells lateral to the RMS were found to co-express both markers (arrowheads). G?I: Cells expressing the radial glia marker S100β (H) with radial glia-like morphologies can be observed co-expressing Tg(olig2:EGFP) (I) in a short stretch ventral to the RMS (arrows). Scale bar = (A?C) 25 μm, (D?F) 25 μm, (D′?F′) 5 μm, (G?I) 25 μm. |
|
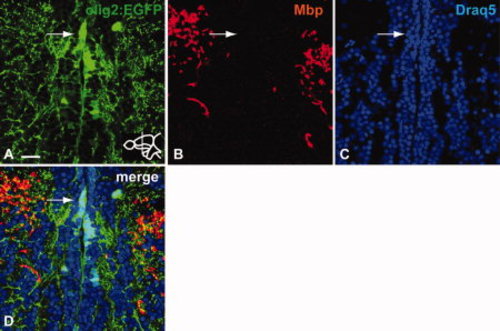
Mbp (B) is not expressed in Tg(olig2:EGFP)-expressing cells (A) of the RMS. Immunohistochemistry on a transverse section of the telencephalon stained with anti-GFP and anti-Mbp antibodies, nuclear staining with Draq5. Arrow indicates lack of Mbp expression in the Tg(olig2:EGFP)-positive domain of the RMS. Scale bar = 25 μm. |