Person
Strähle, Uwe
|
|
Biography and Research Interest
BIOGRAPHY:
Director of the Institute of Biological and Chemical Systems - Biological Information Processing (IBCS-BIP) at the Karlsruhe Institute of Technology (KIT) and Professor of the University of Heidelberg
1985-1988:
Graduate and Ph.D. degrees from German Cancer Research Centre at the University of Heidelberg.
1988-1994:
Postdoctoral employment at the German Cancer Research Centre, the University of Oregon, and the University of Oxford.
1994-2003:
Research Director at CNRS, Group Leader at IGBMC.
University of Strasbourg
2003- :
Karlsruhe Institute of Technology (KIT)
Director of the Institute of Biological and Chemical Systems - Biological Information Processing (previously ITG)
RESEARCH INTERESTS:
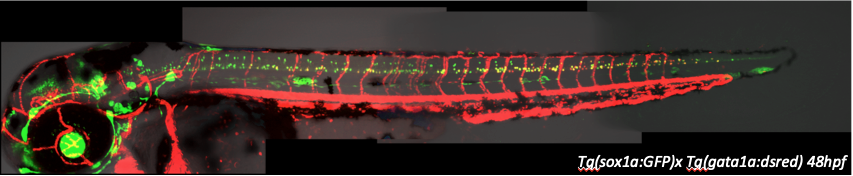
We study the development and regeneration of the vertebrate nervous system and the musculature. Our main goals are to understand the gene networks controlling differentiation and function of the nervous system and musculature, and how these processes are disturbed by environmental toxins. We use genetics and systems biology to model and thereby unravel the underlying regulatory processes. As experimental systems, we employ mostly the zebrafish.
The regulatory architecture of the the zebrafish genome, a systems approach
A key challenge in the post-genome sequencing era is the comprehensive elucidation of the function of the genes and their regulation. The retrieval of genetic information (i.e. expression) is differently regulated depending on cell type and state of the cell. Cross-talk between cells by secretion of signaling molecules is essential for development and body homeostasis. Disturbance of these processes leads to malformations and disease including cancer. In particular, differential transcription into mRNA is a major mechanism controlling the expression of individual genes in development and body homeostasis. Transcription of genes is controlled by specific DNA elements (cis-regulatory elements) that either regulate the access to the DNA via modulating the chromatin structure and/or mediate the recruitment of sequence-specific transcription factors. We refer to all of these genes as transcription regulators (TRs).
The cis-regulatory elements of downstream genes are integration points, at which different signals converge to coordinately regulate gene programs characteristic of the cell states from a pluripotent stem cell to a particular differentiated cell state. A major emphasis of our work is the elucidation and functional characterization of the cis-regulatory architecture of the zebrafish genome. We have undertaken a systematic screen for TRs in the zebrafish genome and determined their expression in the 24 hour old embryo. So far we have mapped the expression pattern of more than 1000 TRs. These patterns will be matched to the temporal and spatial activity of enhancers to derive rules of the regulatory code. In selected structures of the developing central nervous system such as for example the ventral spinal cord, we systematically knock-down TRs and carry out protein/protein and protein/DNA interaction studies to assess the regulatory networks by functional means. The long-term plan is to derive quantitative models of these regulatory processes that integrate all available data in comprehensive and illustrative models that will also be useful to inform and educate the broader public.
Genetics of nervous system and muscle development and maintenance
Motility develops very early in the zebrafish embryo offering a fast assay in screens to identify mutations in genes involved in the development and function of the muscle and the Picture Gallery - Fig. 3embryonic nervous system. We carried out such F3 screens previously and are in the process to clone the affected genes. These zebrafish mutants mimic myopathies and neuropathies of humans. They serve therefore as important models to investigate the molecular mechanisms underlying the corresponding human pathologies.
Development is driven by the controlled differentiation of embryonic stem cells. Stem cells play also crucial roles in the maintenance of tissues in the adult organism. We are interested to elucidate and compare the mechanisms of stem cell maintenance and differentiation in the muscle and nervous system of the embryo and the adult. Zebrafish are well suited for such a research interest as they show even in the adult central nervous system a remarkable ability to regenerate.
The zebrafish embryo as a model for molecular toxicology
We are exposed to a steadily increasing number of different chemicals. A central issue for human health protection is the development of efficient animal models to monitor and to predict the toxic affect of existing and novel compounds. Our aim is to develop the zebrafish as animal model for molecular toxicology. We use microarray technology, transgenesis and genetics to unravel the molecular pathways affected by model toxins. We focus on toxins that affect neural and muscular development. We believe that this work will elucidate the molecular mechanism of toxicity and help to predict the toxicity of novel compounds.
Director of the Institute of Biological and Chemical Systems - Biological Information Processing (IBCS-BIP) at the Karlsruhe Institute of Technology (KIT) and Professor of the University of Heidelberg
1985-1988:
Graduate and Ph.D. degrees from German Cancer Research Centre at the University of Heidelberg.
1988-1994:
Postdoctoral employment at the German Cancer Research Centre, the University of Oregon, and the University of Oxford.
1994-2003:
Research Director at CNRS, Group Leader at IGBMC.
University of Strasbourg
2003- :
Karlsruhe Institute of Technology (KIT)
Director of the Institute of Biological and Chemical Systems - Biological Information Processing (previously ITG)
RESEARCH INTERESTS:
We study the development and regeneration of the vertebrate nervous system and the musculature. Our main goals are to understand the gene networks controlling differentiation and function of the nervous system and musculature, and how these processes are disturbed by environmental toxins. We use genetics and systems biology to model and thereby unravel the underlying regulatory processes. As experimental systems, we employ mostly the zebrafish.
The regulatory architecture of the the zebrafish genome, a systems approach
A key challenge in the post-genome sequencing era is the comprehensive elucidation of the function of the genes and their regulation. The retrieval of genetic information (i.e. expression) is differently regulated depending on cell type and state of the cell. Cross-talk between cells by secretion of signaling molecules is essential for development and body homeostasis. Disturbance of these processes leads to malformations and disease including cancer. In particular, differential transcription into mRNA is a major mechanism controlling the expression of individual genes in development and body homeostasis. Transcription of genes is controlled by specific DNA elements (cis-regulatory elements) that either regulate the access to the DNA via modulating the chromatin structure and/or mediate the recruitment of sequence-specific transcription factors. We refer to all of these genes as transcription regulators (TRs).
The cis-regulatory elements of downstream genes are integration points, at which different signals converge to coordinately regulate gene programs characteristic of the cell states from a pluripotent stem cell to a particular differentiated cell state. A major emphasis of our work is the elucidation and functional characterization of the cis-regulatory architecture of the zebrafish genome. We have undertaken a systematic screen for TRs in the zebrafish genome and determined their expression in the 24 hour old embryo. So far we have mapped the expression pattern of more than 1000 TRs. These patterns will be matched to the temporal and spatial activity of enhancers to derive rules of the regulatory code. In selected structures of the developing central nervous system such as for example the ventral spinal cord, we systematically knock-down TRs and carry out protein/protein and protein/DNA interaction studies to assess the regulatory networks by functional means. The long-term plan is to derive quantitative models of these regulatory processes that integrate all available data in comprehensive and illustrative models that will also be useful to inform and educate the broader public.
Genetics of nervous system and muscle development and maintenance
Motility develops very early in the zebrafish embryo offering a fast assay in screens to identify mutations in genes involved in the development and function of the muscle and the Picture Gallery - Fig. 3embryonic nervous system. We carried out such F3 screens previously and are in the process to clone the affected genes. These zebrafish mutants mimic myopathies and neuropathies of humans. They serve therefore as important models to investigate the molecular mechanisms underlying the corresponding human pathologies.
Development is driven by the controlled differentiation of embryonic stem cells. Stem cells play also crucial roles in the maintenance of tissues in the adult organism. We are interested to elucidate and compare the mechanisms of stem cell maintenance and differentiation in the muscle and nervous system of the embryo and the adult. Zebrafish are well suited for such a research interest as they show even in the adult central nervous system a remarkable ability to regenerate.
The zebrafish embryo as a model for molecular toxicology
We are exposed to a steadily increasing number of different chemicals. A central issue for human health protection is the development of efficient animal models to monitor and to predict the toxic affect of existing and novel compounds. Our aim is to develop the zebrafish as animal model for molecular toxicology. We use microarray technology, transgenesis and genetics to unravel the molecular pathways affected by model toxins. We focus on toxins that affect neural and muscular development. We believe that this work will elucidate the molecular mechanism of toxicity and help to predict the toxicity of novel compounds.
Non-Zebrafish Publications
Str�hle U, Boshart M, Klock G, Stewart F, Sch�tz G (1989). Glucocorticoid- and progesterone-specific effects are determined by differential expression of the respective hormone receptors. Nature.339(6226):629-32.
Str�hle U, Schmid W, Sch�tz G (1988).Synergistic action of the glucocorticoid receptor with transcription factors.EMBO J.7(11):3389-95
Str�hle U, Klock G, Sch�tz G (1987). A DNA sequence of 15 base pairs is sufficient to mediate both glucocorticoid and progesterone induction of gene expression. Proc Natl Acad Sci U S A.84(22):7871-5
