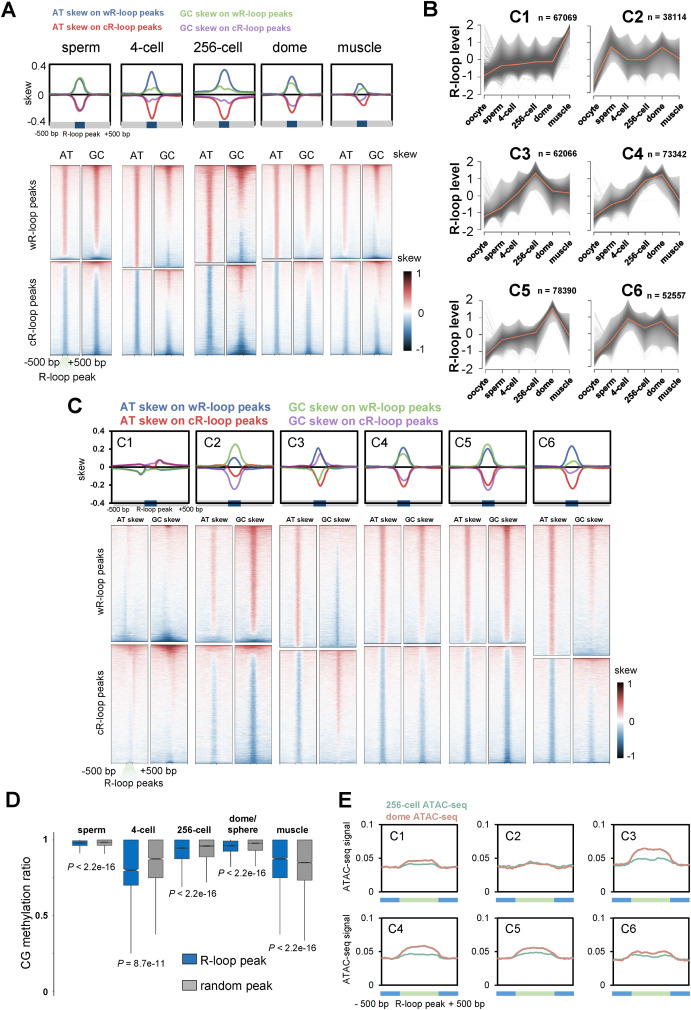
Fig. 4 Characteristics of R-loop dynamics during early development. A. Metaplots (upper) and heatmaps (lower) of AT or GC skew around R-loop peaks of sperm, 4-cell, 256-cell, dome and muscle groups. B. Fuzzy cluster analysis of R-loop signals by using Mfuzz. Line plots show ULI-ssDRIP-seq signals, with individual loci (grey lines) and values of the cluster center (orange line). The count of R-loop peaks from each cluster is shown as “n”. R-loop peak locations of each cluster are shown in Table S4. C. Metaplots (upper) and heatmaps (lower) of AT or GC skew around R-loop peaks of clusters 1 to 6 shown in Fig. 4B. D. Box plot of CG methylation levels (SRP020008) on R-loop peaks (blue) or random peaks (grey) in sperm, muscle, and 4-cell, 256-cell, dome/sphere stage embryos. As sphere-stage is very close to dome stage, the CG methylation pattern at sphere stage is analyzed with R-loop peaks at dome stage. P values were calculated using Mann-Whitney U test. E. Metaplots of ATAC-seq signals (GSE101779) around six R-loop clusters (Fig. 4B) in zebrafish embryos at 256-cell or dome stage.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Cell Insight