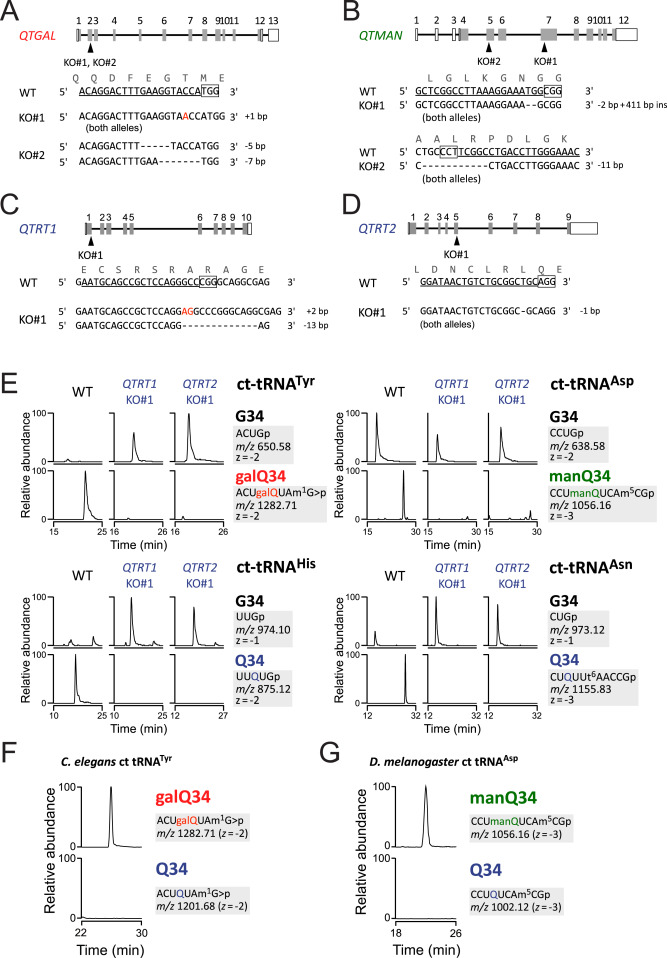
Fig. S2 Genetic characterization of Q and Q-glycosylation in human cells, related to Figure 4 (A–D) Schematic diagram of human QTRT1 (A), QTRT2 (B), QTGAL (C), and QTMAN (D) genes with sgRNA sequences. Shaded and open boxes indicate coding and untranslated regions of the genes, respectively. Lines indicate introns. Mutation sites in sgRNA-targeted exons are indicated by arrowheads. The sgRNA-targeted sequences are underlined and protospacer adjacent motif (PAM) sequences are boxed. Nucleotide insertion and deletion are indicated by red letters and dashed lines, respectively. (E) XICs showing the RNase T1-digested fragments containing position 34 of cytoplasmic tRNAs for Tyr, Asp, His, and Asn isolated from WT HEK293T (left), QTRT1 KO (middle), and QTRT2 KO (right). Signal intensities were normalized against a unique internal fragment of respective tRNAs. t6A, N6-threonylcarbamoyladenosine. (F and G) Analyses of glycosyl-Qs in other animals. XICs showing the galQ-containing fragment of C. elegans cytoplasmic tRNATyr (F), and the manQ-containing fragment of D. melanogaster cytoplasmic tRNAAsp (G).
Reprinted from Cell, 186(25), Zhao, X., Ma, D., Ishiguro, K., Saito, H., Akichika, S., Matsuzawa, I., Mito, M., Irie, T., Ishibashi, K., Wakabayashi, K., Sakaguchi, Y., Yokoyama, T., Mishima, Y., Shirouzu, M., Iwasaki, S., Suzuki, T., Suzuki, T., Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth, 5517-5535.e24, Copyright (2023) with permission from Elsevier. Full text @ Cell