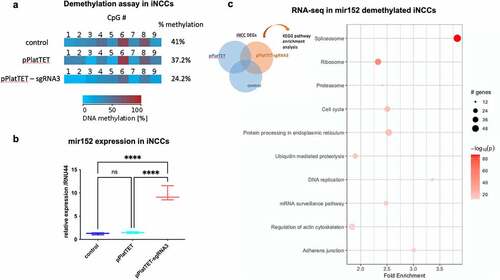
Fig. 2
Figure 2. DNA methylation changes at mir152 DMRs results in mir152 expression changes and affects Spliceosomal, Ribosomal, and Adherens Junctions pathways. (a) A Cas9-based approach for target demethylation using the vector pPlatTET and mir152 specific single-guide RNA sequences (sgRNA3) for mir152 DMR in iNCC. sgRNA-3 efficiently reduces mir152 methylation in comparison to the empty vector transfection (pPlatTET) and non-transfected cells (control). Total percentage of methylation is represented with values at the right. (b) Targeted mir152 DNA hypomethylation by pPlatTET-sgRNA-3 (modified Cas9 coupled with the catalytic domain of TET1) induces significant mir152 overexpression in induced neural crest cells (iNCCs) revealed by RT-qPCR. Relative expression to endogenous control RNU44. Mann?Whitney?s test. (c) Enriched KEGG pathways found in RNA-seq analysis in mir152 demethylated iNCCs (pPlatTET-sgRNA3), overexpressing mir152. Higher fold enrichments are observed in Spliceosome and Adherens Junctions DEGs.