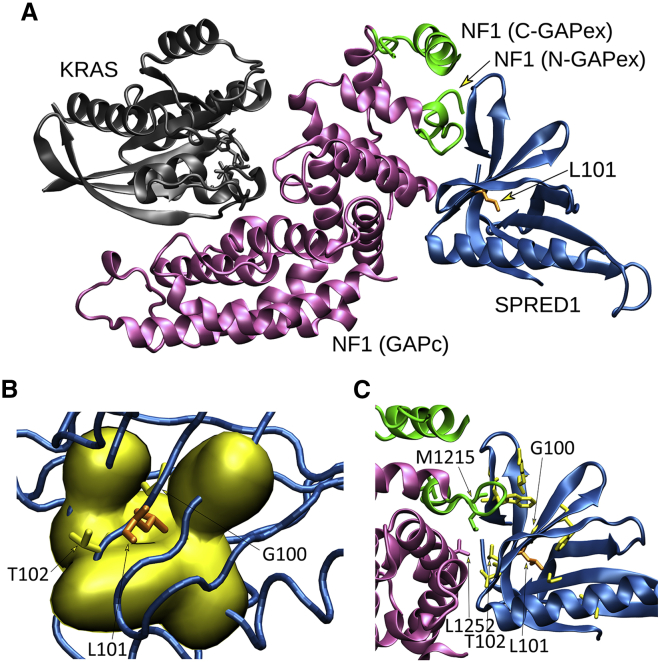
Fig. 3
Structural impact of the p.Leu100Pro substitution in SPRED2
(A) The crystallographic complex of neurofibromin (NF1) GRD (GAPc, magenta) with the EVH1 domain of SPRED1 (blue) and GTP-bound KRAS (gray) is shown. The N- and C-terminal GAPex stretches encompassing GAPc are shown in green. The lateral chain of Leu101 (L101) (corresponding to Leu100 in SPRED2) is shown (orange).
(B) Surface visualization (Gaussian density surface) of the hydrophobic residues close to Leu101 (i.e., Ala19, Val21, Val42, Trp92, Phe103, and Phe112) is shown in yellow. Lateral chains of Leu101 and adjacent residues, Gly100 (G100) and Thr102 (T102), mutated in LGSS, are shown.
(C) Enlarged view of the neurofibromin and SPRED1 interacting surfaces. Relevant residues are indicated using the single-letter amino acid code. Lateral chains of SPRED1 residues (Arg24, Gly30, Trp31, Val44, Gly62, Thr88, Trp92, Gly100, and Thr102) mutated in LGSS are shown (yellow), together with Leu101 (orange). Of note, Thr102 interacts with both GAPc (Leu1252) and GAPex (Met1215).