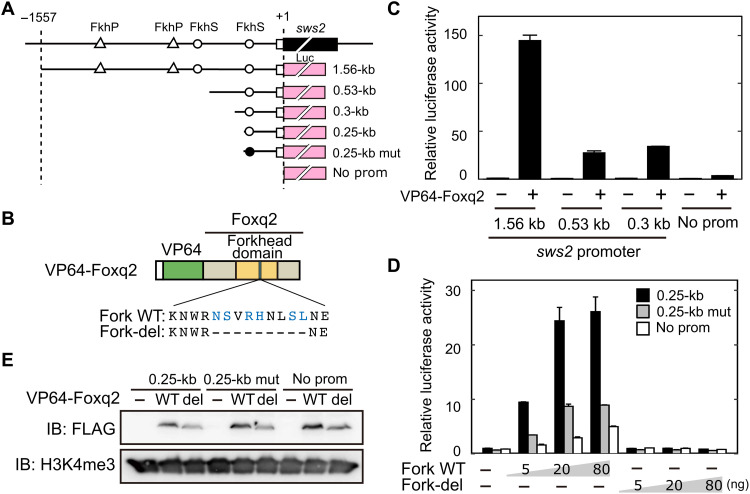
Fig. 5
(A) Schematic structure of the promoter-luciferase reporter constructs for zebrafish sws2. The coding (solid box) and the untranslated (open box) regions of the first exon are indicated. The translational start site is marked as +1. The forkhead binding motif (ACAACA) and its mutant (GTGGTG) are marked as open and closed circles, respectively. See also fig. S6. (B) Schematic representation of VP64-Foxq2 protein. The Foxq2 mutant protein (Fork-del) lacks the canonical Fox base?contacting residues in the forkhead domain. These critical amino acid residues are highlighted in blue according to the previous paper (30). (C and D) Transcriptional assay in human embryonic kidney 293T17 cells using the luciferase reporter containing the promoter region of zebrafish sws2 (means ± SD, n = 2). The luciferase activity derived from the firefly luciferase reporter was normalized to that from the Renilla luciferase reporter. (C) The luciferase activity in each condition was normalized to the mean value obtained from the cells transfected with both empty vector and 1.56-kb sws2 reporter vector (leftmost bar). VP64-Foxq2 expression plasmid (100 ng) was used for the transfection. (D) The amount of VP64-Foxq2 expression plasmid used is indicated. The luciferase activity in each condition was normalized to the mean value obtained from the cells transfected with both the empty expression vector and the 0.25-kb sws2 (0.25 kb) reporter vector (leftmost bar). The slight up-regulation of the luciferase expression in a reporter with no sws2 promoter might be due to an experimental artifact, such as VP64-Foxq2?dependent regulation through the reporter vector-backbone sequence(s) and/or activation of gene(s) regulating the reporter gene expression. (E) Protein expression of VP64-Foxq2 (WT) and its mutant protein (Fork-del) in the cells transfected with 80 ng of the plasmid. The antibody H3K4me3 is served as a loading control. IB, immunoblot.