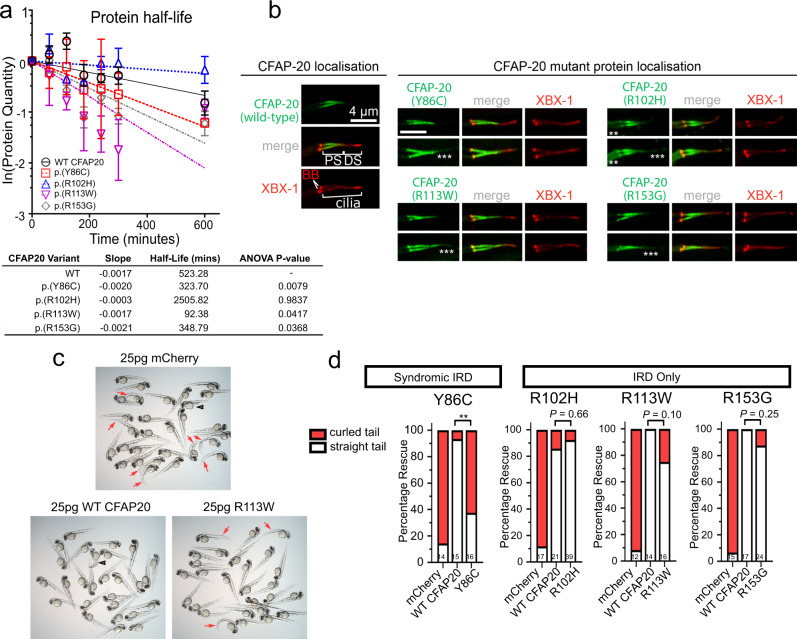
Fig. 6
a Half life calculations assuming first-order decay (ln([relative protein]t = x/[relative protein]t = 0)). Lysates from transfected HEK293T cells revealed a significant (one-way ANOVA) decay for all variants, except for p.(Arg102His), compared with Wildtype (WT) protein (half-life of ~523 min). The fastest decay was variant p.(Arg113Trp) with a half-life of ~93 min (n = 3; error bars = SEM). b In C. elegans, human CFAP20 patient variants mislocalise in comparison to wildtype CFAP20. The GFP-tagged CFAP20 proteins bearing patient mutations concentrate mostly to the proximal segment (PS) of the axoneme, which is similar to wild-type CFAP20 protein expressed in C. elegans. However, patient variants also mislocalise (asterisks) in the distal segment (DS) and/or the dendritic region to the left of the phasmid cilia (especially p.(Arg102His)). XBX-1 is an IFT reporter that marks the basal body (BB) and entire axoneme. c Wildtype human CFAP20 mRNA rescues zebrafish cfap20 mutants, as compared to control mRNA (mCherry) or CFAP20 mRNA bearing patient variants. A representative cfap20+/? incross is displayed, injected with WT/patient variant (p.(Arg113Trp)) CFAP20, or control mRNA. Mendelian ratios of 25% body axis curvature (red arrows) were expected where treatments failed to rescue the cfap20?/? homozygotes. Larvae displaying mRNA toxicity phenotypes (i.e. oedema/dorsalization) occur rarely (black arrowhead). d The frequency of rescued (straight tail) and not rescued (curled tail) cfap20?/? homozygotes were quantified for each missense human variant. p.(Tyr86Cys), which is associated with syndromic inherited retinal disease (IRD), failed to rescue body curvature. Conversely, the p.(Arg102His), p.(Arg113Trp), and p.(Arg153Gly) variants, that we associated with non-syndromic IRD, are not significantly different from WT CFAP20 mRNA (Fisher?s exact test; p.(Tyr86Cys) P = 0.0021; sample sizes, i.e. the number of cfap20?/? homozygous mutant larvae, are indicated at the bottom of each bar). See also Supplementary Fig. 8. Source data are provided as a Source Data file.