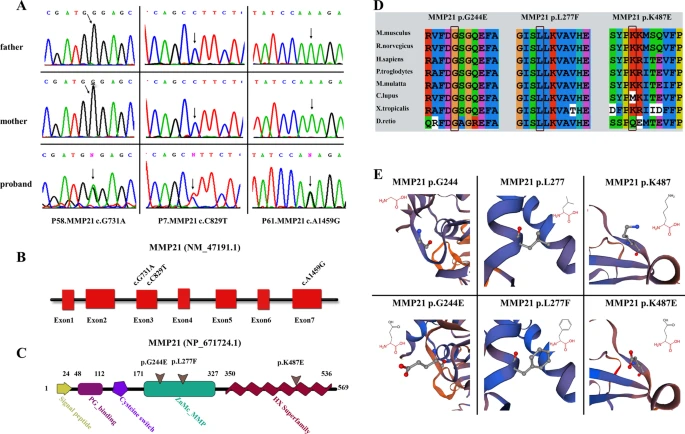
Fig. 1
Validation and in silico analysis of MMP21 variants found in HTX patients with complex CHDs. A Sanger sequencing chromatograms of three heterozygous missense MMP21 variants in patients with HTX and complex CHDs and the corresponding normal sequences in related parents. Arrows represent heterozygous nucleotide changes. B, C Schematic diagrams of MMP21 gene and protein showing the location of the identified MMP21 variants. The base substitutions are in exon3 and exon7, respectively (B). The amino acids consist of the domains of the ZnMC_MMP and HX superfamily (C). D Clustal X alignment of amino acid mutations in MMP21 among different species indicates that all mutations are highly conserved in vertebrates. E Schematic diagram of the three-dimensional structure and corresponding amino acid residues at the mutation sites of WT (left panel) and mutant (right panel) MMP21 proteins that were reconstructed by SWISS-MODEL. Amino acid substitutions in the variants p.G244E and p.K487E resulted in changes in acidity or basicity, which may affect the function of the mutant domain. The amino acid residue in the original position of the variant p.L277F was replaced by a residue that occupies a larger space volume, which may affect the stability of the protein spatial structure