Fig. 3
Image
Figure Caption
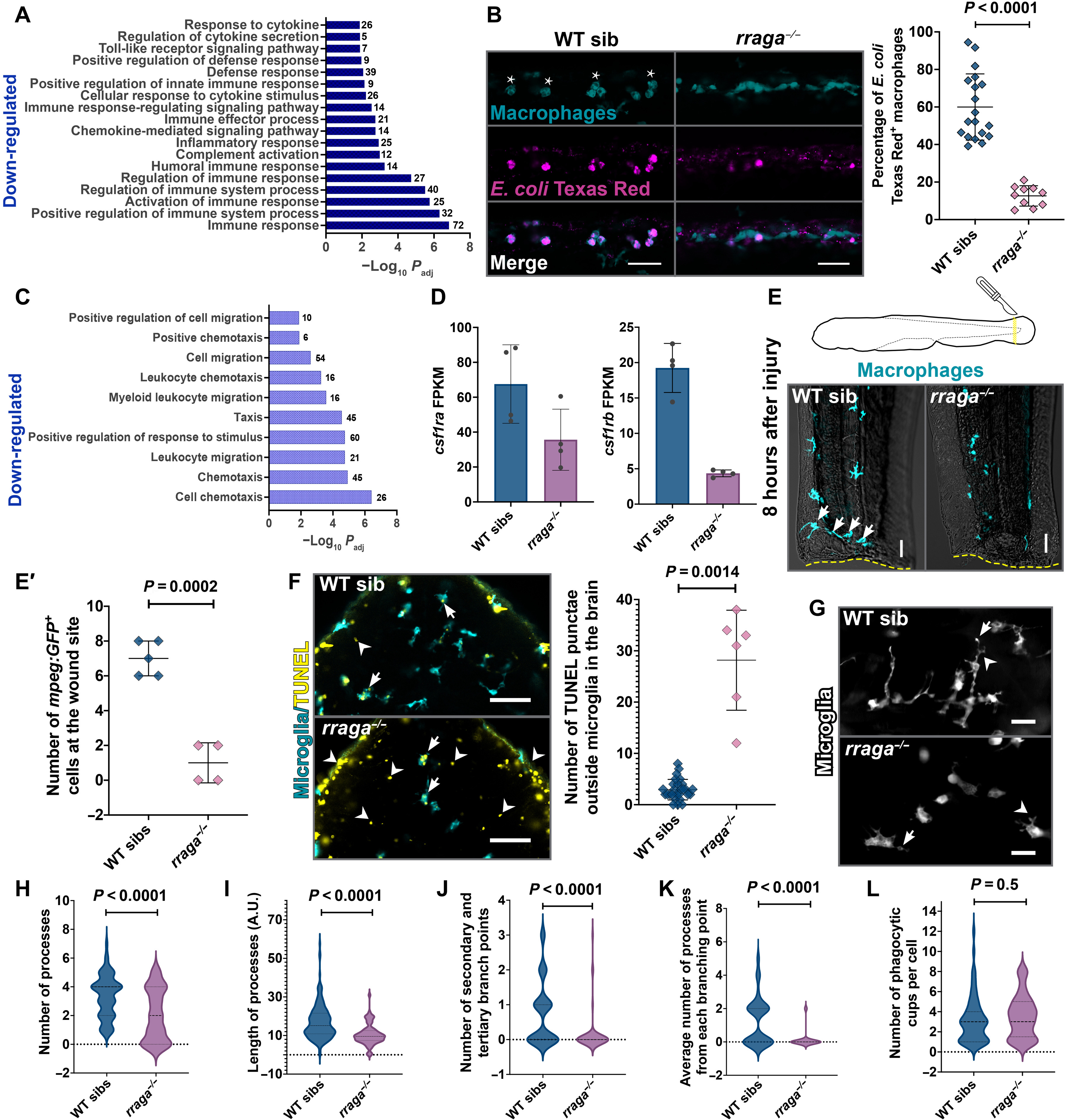
Fig. 3. Macrophages and microglia in rraga mutants show defective clearance of debris.
(A) Bar graph showing GO terms related to immune signaling significantly down-regulated in macrophages from rraga mutants. (B) Injection of E. coli Texas Red in rraga mutants and siblings at 4 dpf and corresponding quantification. Wild-type macrophages responding to E. coli become activated and display amoeboid morphology (asterisks). Scale bars, 50 ?m. (C) GO terms related to migration/taxis significantly down-regulated in macrophages from rraga mutants. (D) Graphs showing fragments per kilobase of exon per million mapped reads (FPKM) values of csf1ra and csf1rb transcripts in macrophages from rraga mutants; each point represents FPKM value from a single RNA-seq biological replicate. FPKM, fragments per kilobase of exon per million mapped reads. (E and E?) Macrophage response to tail injury at 4 dpf. Arrows in the wild-type image show macrophages at the wound site (yellow dotted line). Scale bars, 20 ?m. (F) TUNEL assay and quantification on rraga mutants and their wild-type siblings. Arrows indicate TUNEL+ microglia, and arrowheads indicate TUNEL signal outside microglia. Scale bars, 50 ?m. (G to L) Time-lapse imaging and analysis of cellular characteristics of microglia at 5 dpf. (G) Images of rraga mutants and their siblings, with arrows showing phagocytic cup formation and arrowheads showing branching points. Scale bars, 20 ?m. All graphs show mean + SD. Significance in (B) was determined using nonparametric Mann-Whitney U test; significance in (E?), (F), and (H) to (L) was determined using parametric unpaired t test. The number of animals analyzed for each experiment is listed in table S1; all the panels are representative of at least two independent experiments.
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Sci Adv