Fig. 2
Image
Figure Caption
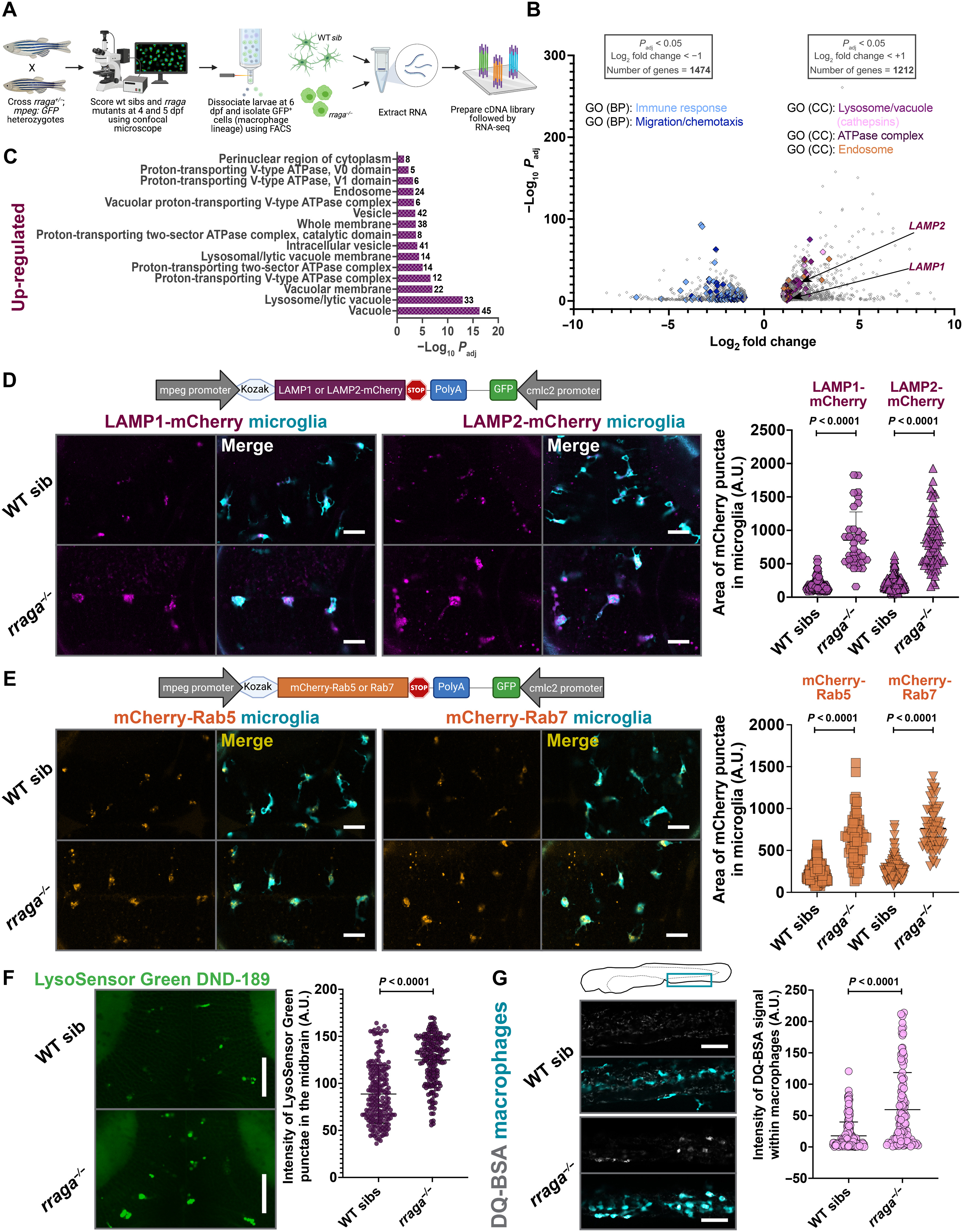
Fig. 2. Lysosomal genes are significantly up-regulated and endolysosomes are expanded in macrophages isolated from rraga mutants.
(A) Experimental schematic for RNA-seq. Macrophages were isolated using FACS from N = 90 larvae for each biological replicate RNA sample from rraga mutants or wild-type siblings. Four biological replicate samples of each genotype were processed for RNA-seq. (B) Volcano plot showing significantly differentially up-regulated and down-regulated genes in the macrophages of rraga mutants relative to wild-type siblings. (C) GO (Cellular Component) enrichment analysis showing an up-regulation of lysosome-associated terms in macrophages from rraga mutants. (D) Imaging and quantification of LAMP1 or LAMP2-mCherry transgene expression in rraga mutants and wild-type siblings at 4 dpf. Scale bars, 20 ?m. (E) Imaging and quantification of mCherry-Rab5 or Rab7 transgene expression in rraga mutants and wild-type siblings at 4 dpf. Scale bars, 20 ?m. (F) LysoSensor Green labeling and quantification of LysoSensor Green intensity at 4 dpf. Scale bars, 50 ?m. Graphs in (D) to (F) show mean + SD; significance was determined using parametric unpaired t test. (G) DQ-BSA labeling at 4 dpf and quantification of DQ-BSA intensity inside macrophages. Scale bars, 50 ?m. Graph shows mean + SD; significance was determined using nonparametric Mann-Whitney U test. The number of animals analyzed for each experiment is listed in table S1; all the panels (except RNA-seq) are representative of at least two independent experiments. A.U., arbitrary units.
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Sci Adv