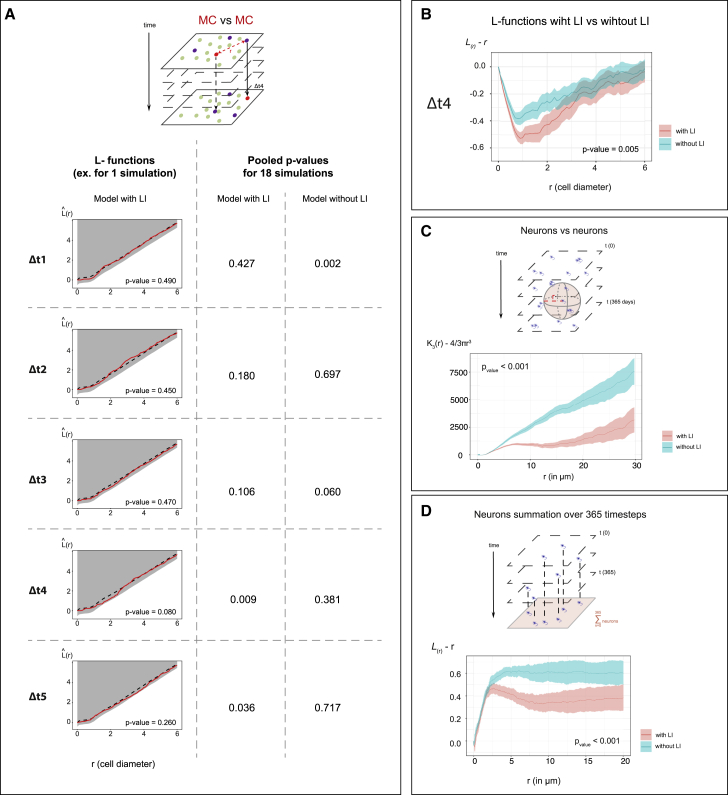
Figure 7
The NSC lattice model reproduces the spatiotemporal correlations of NSC fate decisions and shows their impact on long-term neuronal distribution
(A) Top: MC positions at any time t in the lattice model are compared using point pattern statistics with MC positions at t+Δt. Bottom left: Besag’s
(B) Compared
(C) Compared spatial interaction between output neurons in a model with (pink) and without (blue) LI, using a 3D Ripley’s
(D) Similar comparison (to C) in 2D, using the
(B)–(D) show centered summary functions,